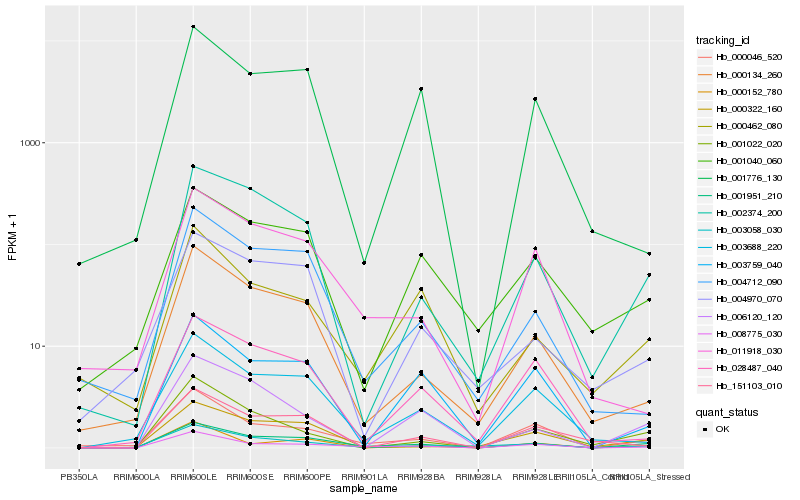
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008775_030 |
0.1665418985 |
- |
- |
PREDICTED: ferric reduction oxidase 7, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_002374_200 |
0.1667479777 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 4 |
Hb_000046_520 |
0.1731136166 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003058_030 |
0.1806688791 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000134_260 |
0.1863455788 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_006120_120 |
0.1868049668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000322_160 |
0.1886332849 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2-like [Jatropha curcas] |
| 9 |
Hb_001040_060 |
0.1916952598 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 10 |
Hb_003759_040 |
0.1922361279 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 11 |
Hb_003688_220 |
0.1959441929 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_000152_780 |
0.1981336656 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 13 |
Hb_000462_080 |
0.2013150075 |
- |
- |
PREDICTED: quercetin 3-O-methyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_004712_090 |
0.2037131452 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_004970_070 |
0.2050710138 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 16 |
Hb_001022_020 |
0.2056094493 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 17 |
Hb_011918_030 |
0.2160931953 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 18 |
Hb_028487_040 |
0.2161763681 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 19 |
Hb_151103_010 |
0.2165586297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001776_130 |
0.2175186531 |
- |
- |
metallothionein [Hevea brasiliensis] |