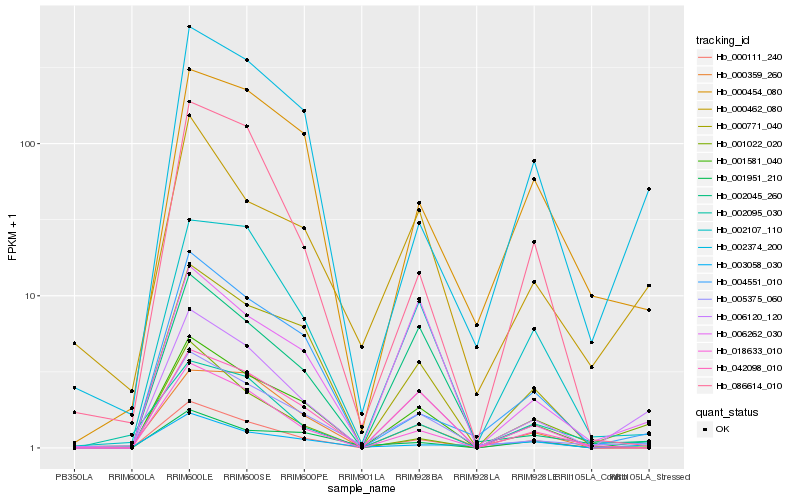
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006120_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001581_040 |
0.1652385489 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_005375_060 |
0.1686946643 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 4 |
Hb_000111_240 |
0.1742044509 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 5 |
Hb_002374_200 |
0.1742864434 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 6 |
Hb_001022_020 |
0.1848623631 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 7 |
Hb_001951_210 |
0.1868049668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002107_110 |
0.1874239924 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_018633_010 |
0.1879057869 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000462_080 |
0.1924557595 |
- |
- |
PREDICTED: quercetin 3-O-methyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_042098_010 |
0.2004205479 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 12 |
Hb_000771_040 |
0.2017234483 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 13 |
Hb_006262_030 |
0.2074263709 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002045_260 |
0.2075051278 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 15 |
Hb_004551_010 |
0.2076229799 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 16 |
Hb_002095_030 |
0.2080724604 |
- |
- |
hypothetical protein MIMGU_mgv1a021906mg, partial [Erythranthe guttata] |
| 17 |
Hb_000359_260 |
0.2104655376 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 18 |
Hb_086614_010 |
0.2129653086 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_003058_030 |
0.2135542652 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000454_080 |
0.2151591957 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |