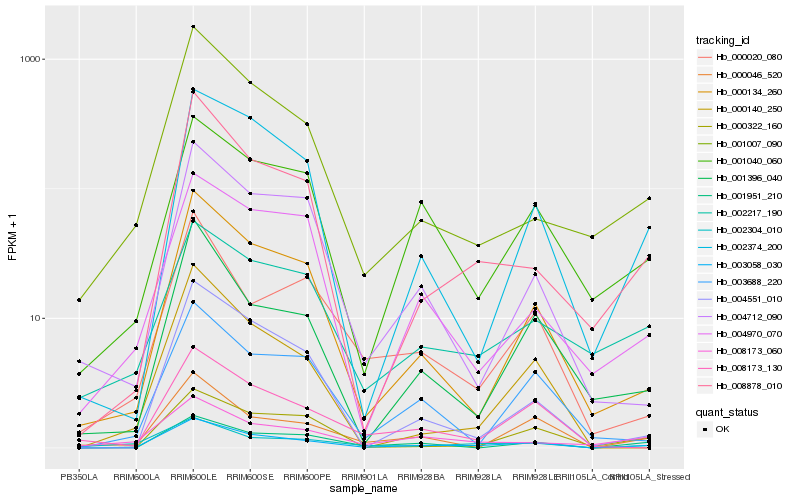
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003058_030 |
0.0 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000134_260 |
0.1493093442 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_008878_010 |
0.1594036668 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 4 |
Hb_002374_200 |
0.1659270505 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 5 |
Hb_001007_090 |
0.1700255815 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 6 |
Hb_001040_060 |
0.1712593731 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 7 |
Hb_001396_040 |
0.1786346364 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 8 |
Hb_002304_010 |
0.1787131607 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 9 |
Hb_000322_160 |
0.1793747807 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2-like [Jatropha curcas] |
| 10 |
Hb_002217_190 |
0.1803711621 |
- |
- |
invertase [Hevea brasiliensis] |
| 11 |
Hb_001951_210 |
0.1806688791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004551_010 |
0.1825226728 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 13 |
Hb_000046_520 |
0.1825333168 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003688_220 |
0.1872631943 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_004970_070 |
0.1873280716 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 16 |
Hb_000020_080 |
0.19086756 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 17 |
Hb_000140_250 |
0.1917936292 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 18 |
Hb_008173_130 |
0.192234765 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008173_060 |
0.1929353147 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_004712_090 |
0.1946787358 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |