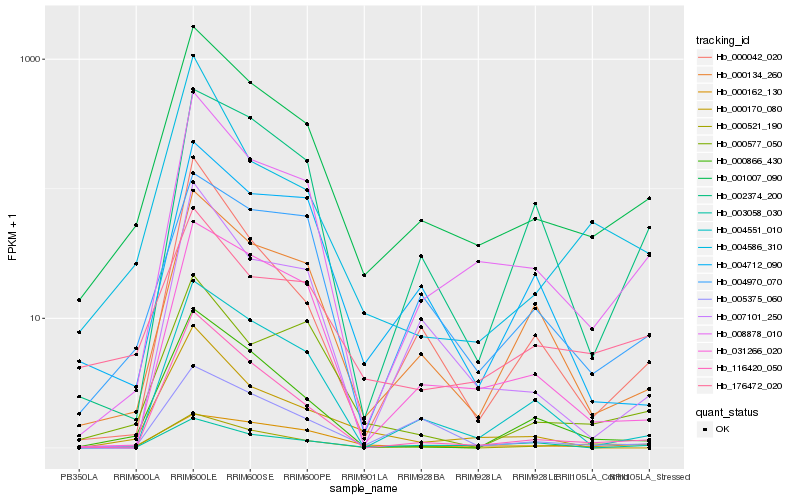
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001007_090 |
0.0 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 2 |
Hb_008878_010 |
0.1197168217 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 3 |
Hb_000521_190 |
0.1235015339 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_25500 [Jatropha curcas] |
| 4 |
Hb_000134_260 |
0.1482334695 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_176472_020 |
0.1533655452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000866_430 |
0.1572806919 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 7 |
Hb_116420_050 |
0.1594135434 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 8 |
Hb_031266_020 |
0.1635052629 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004970_070 |
0.1686381847 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 10 |
Hb_003058_030 |
0.1700255815 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000162_130 |
0.1700529047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004551_010 |
0.1708224071 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 13 |
Hb_000042_020 |
0.1750517644 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_004586_310 |
0.1759755955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000577_050 |
0.1786045773 |
- |
- |
- |
| 16 |
Hb_002374_200 |
0.1792303439 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 17 |
Hb_004712_090 |
0.1813713215 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 18 |
Hb_007101_250 |
0.1820975363 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 19 |
Hb_005375_060 |
0.182187938 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 20 |
Hb_000170_080 |
0.1845081316 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |