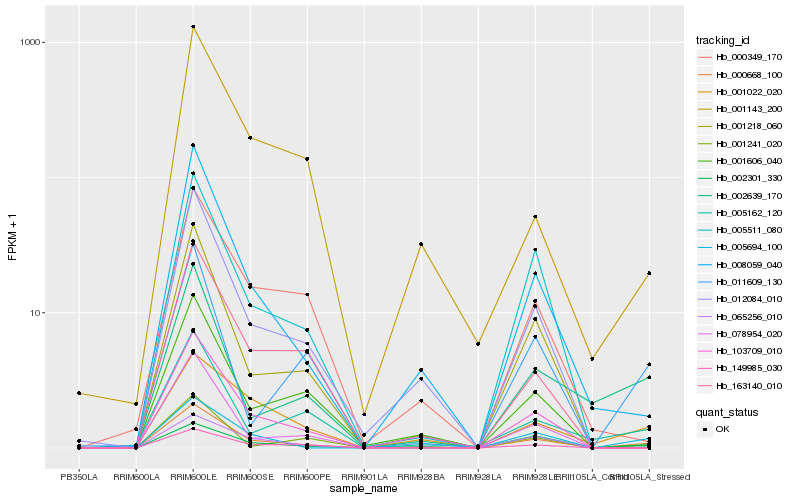
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_100 |
0.0 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 2 |
Hb_005694_100 |
0.1543860438 |
- |
- |
PREDICTED: calcium-binding protein KIC-like [Populus euphratica] |
| 3 |
Hb_008059_040 |
0.1844936097 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 4 |
Hb_005511_080 |
0.1934702094 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 5 |
Hb_001022_020 |
0.1945440523 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 6 |
Hb_001241_020 |
0.1961342654 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 7 |
Hb_001218_060 |
0.1966537647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002301_330 |
0.1987520428 |
- |
- |
PREDICTED: late embryogenesis abundant protein, group 3-like [Jatropha curcas] |
| 9 |
Hb_078954_020 |
0.1992184984 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 10 |
Hb_002639_170 |
0.1994218521 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 11 |
Hb_011609_130 |
0.2001681337 |
- |
- |
- |
| 12 |
Hb_103709_010 |
0.201201908 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 13 |
Hb_012084_010 |
0.2048395362 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 14 |
Hb_005162_120 |
0.2049842121 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 15 |
Hb_001606_040 |
0.2067534714 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 16 |
Hb_163140_010 |
0.2073941541 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 17 |
Hb_065256_010 |
0.2128978558 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 18 |
Hb_001143_200 |
0.2180920384 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 19 |
Hb_149985_030 |
0.2187745647 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 20 |
Hb_000349_170 |
0.2190612086 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |