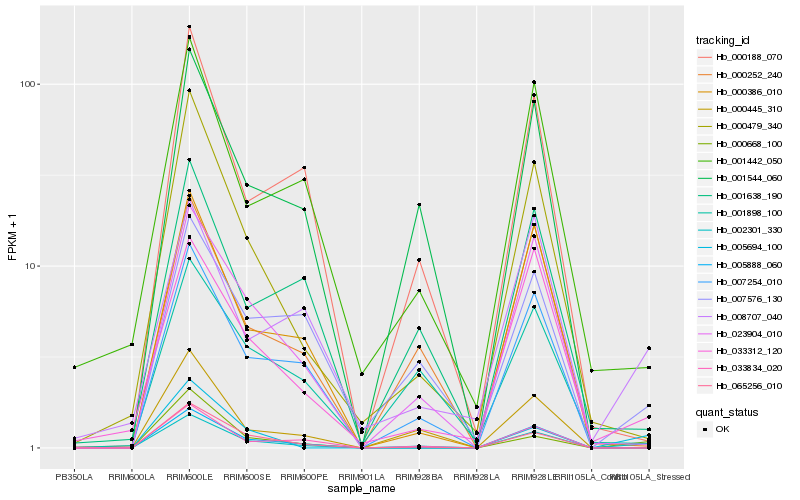
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_330 |
0.0 |
- |
- |
PREDICTED: late embryogenesis abundant protein, group 3-like [Jatropha curcas] |
| 2 |
Hb_023904_010 |
0.1878410769 |
- |
- |
- |
| 3 |
Hb_000479_340 |
0.1937973479 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 4 |
Hb_007576_130 |
0.1943799534 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 5 |
Hb_033312_120 |
0.1950180621 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000668_100 |
0.1987520428 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 7 |
Hb_007254_010 |
0.1991693117 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 8 |
Hb_000445_310 |
0.201373841 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 9 |
Hb_065256_010 |
0.2024368936 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 10 |
Hb_005694_100 |
0.2046338349 |
- |
- |
PREDICTED: calcium-binding protein KIC-like [Populus euphratica] |
| 11 |
Hb_000252_240 |
0.2058634995 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |
| 12 |
Hb_033834_020 |
0.2058756032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008707_040 |
0.2071038677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001544_060 |
0.2081095351 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005888_060 |
0.2103476842 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 16 |
Hb_000188_070 |
0.212566883 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 17 |
Hb_001638_190 |
0.2152180071 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000386_010 |
0.2160844966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001442_050 |
0.2165381966 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001898_100 |
0.2171150707 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |