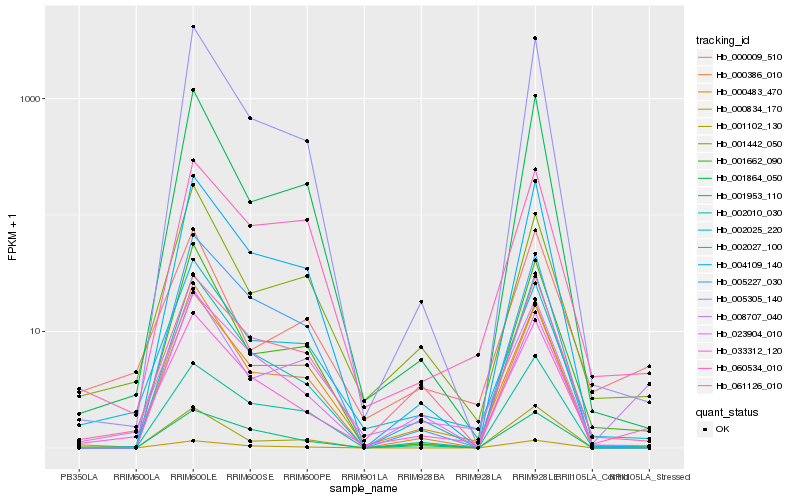
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_120 |
0.0 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002027_100 |
0.1334019256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002025_220 |
0.1340732196 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_061126_010 |
0.1368540864 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_000483_470 |
0.141401549 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_023904_010 |
0.142302959 |
- |
- |
- |
| 7 |
Hb_005305_140 |
0.1429309522 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_001442_050 |
0.1509817043 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004109_140 |
0.1512805579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008707_040 |
0.1558110739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000834_170 |
0.1580336572 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 12 |
Hb_001662_090 |
0.1582539735 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002010_030 |
0.1584607751 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_000009_510 |
0.1584835627 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 15 |
Hb_001953_110 |
0.1592613718 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 16 |
Hb_001102_130 |
0.1600644928 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 17 |
Hb_000386_010 |
0.1636855668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005227_030 |
0.1643957216 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 19 |
Hb_060534_010 |
0.1651780636 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 20 |
Hb_001864_050 |
0.1660207937 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |