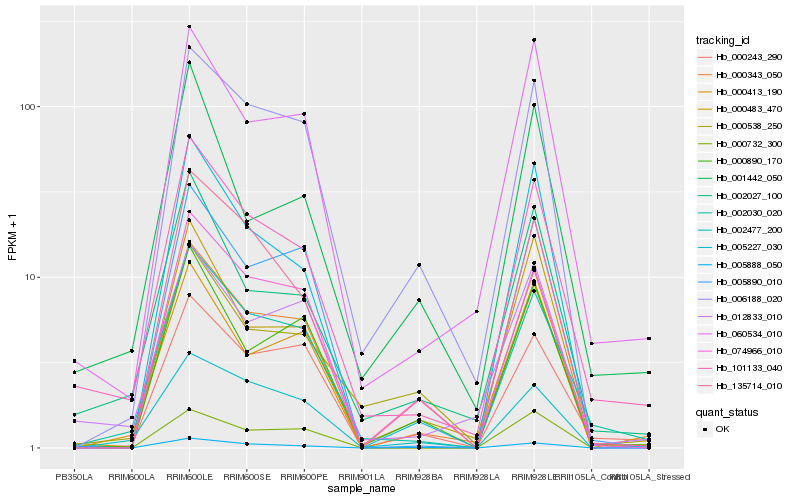
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101133_040 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_002030_020 |
0.0787141669 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 3 |
Hb_002027_100 |
0.1134655389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_060534_010 |
0.1205122814 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 5 |
Hb_000343_050 |
0.1270215849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012833_010 |
0.1365767256 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 7 |
Hb_135714_010 |
0.141645208 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 8 |
Hb_000243_290 |
0.1423573298 |
- |
- |
plastidic aldolase family protein [Populus trichocarpa] |
| 9 |
Hb_005227_030 |
0.1453235134 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000538_250 |
0.1466331852 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 [Jatropha curcas] |
| 11 |
Hb_006188_020 |
0.1491423583 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 12 |
Hb_005888_050 |
0.1498449854 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_002477_200 |
0.1510449255 |
- |
- |
PREDICTED: uncharacterized protein LOC105631678 [Jatropha curcas] |
| 14 |
Hb_000483_470 |
0.1519499778 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001442_050 |
0.1536744837 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 16 |
Hb_074966_010 |
0.1560682276 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 17 |
Hb_000890_170 |
0.1565410383 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 18 |
Hb_000413_190 |
0.1583392338 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 19 |
Hb_005890_010 |
0.1591416644 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 20 |
Hb_000732_300 |
0.161170106 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |