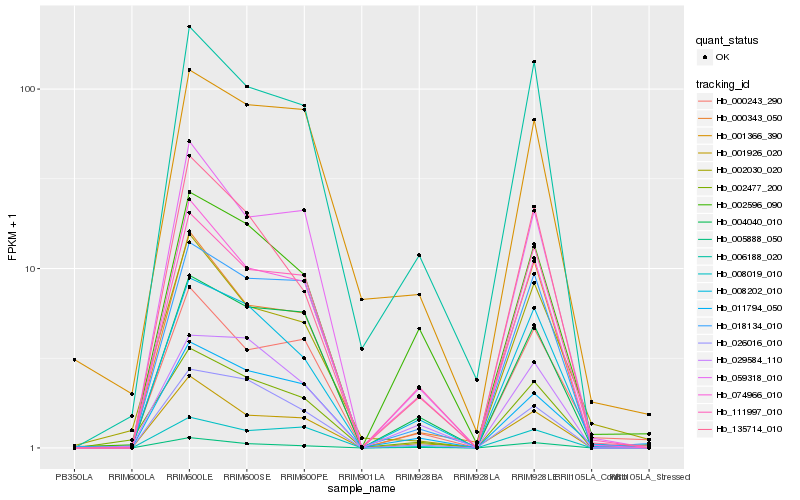
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631678 [Jatropha curcas] |
| 2 |
Hb_008202_010 |
0.1203063553 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_006188_020 |
0.1208278672 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 4 |
Hb_074966_010 |
0.1228939134 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 5 |
Hb_002030_020 |
0.1257492759 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 6 |
Hb_018134_010 |
0.1258085908 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 7 |
Hb_111997_010 |
0.1335695369 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 8 |
Hb_004040_010 |
0.1349408555 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 9 |
Hb_026016_010 |
0.1361992386 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 10 |
Hb_000243_290 |
0.1370895786 |
- |
- |
plastidic aldolase family protein [Populus trichocarpa] |
| 11 |
Hb_005888_050 |
0.1381483401 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_059318_010 |
0.1390322471 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 13 |
Hb_029584_110 |
0.1395258726 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 14 |
Hb_001926_020 |
0.1414914209 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 15 |
Hb_000343_050 |
0.141910981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_135714_010 |
0.1443679213 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 17 |
Hb_002596_090 |
0.1455487458 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001366_390 |
0.14755747 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 19 |
Hb_011794_050 |
0.1483742879 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_008019_010 |
0.1488377092 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |