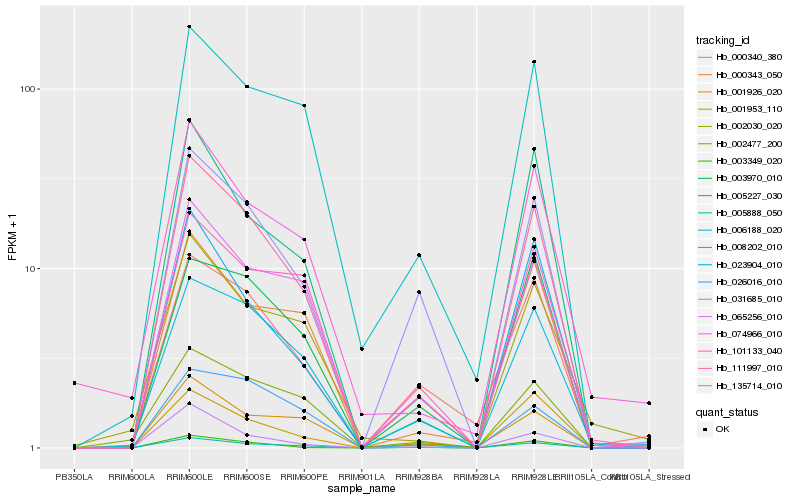
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135714_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 2 |
Hb_005888_050 |
0.0612456657 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_008202_010 |
0.0946909111 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_074966_010 |
0.1125446112 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 5 |
Hb_005227_030 |
0.1199749398 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 6 |
Hb_031685_010 |
0.1241650389 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000343_050 |
0.1241756582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_023904_010 |
0.1285223902 |
- |
- |
- |
| 9 |
Hb_006188_020 |
0.1313422653 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 10 |
Hb_001926_020 |
0.131683926 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 11 |
Hb_001953_110 |
0.1333555095 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 12 |
Hb_111997_010 |
0.1387788162 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 13 |
Hb_003349_020 |
0.1404313524 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_101133_040 |
0.141645208 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_003970_010 |
0.1423528344 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_000340_380 |
0.1443515692 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 17 |
Hb_002477_200 |
0.1443679213 |
- |
- |
PREDICTED: uncharacterized protein LOC105631678 [Jatropha curcas] |
| 18 |
Hb_002030_020 |
0.146849774 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 19 |
Hb_065256_010 |
0.1469924738 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 20 |
Hb_026016_010 |
0.1470786305 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |