| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
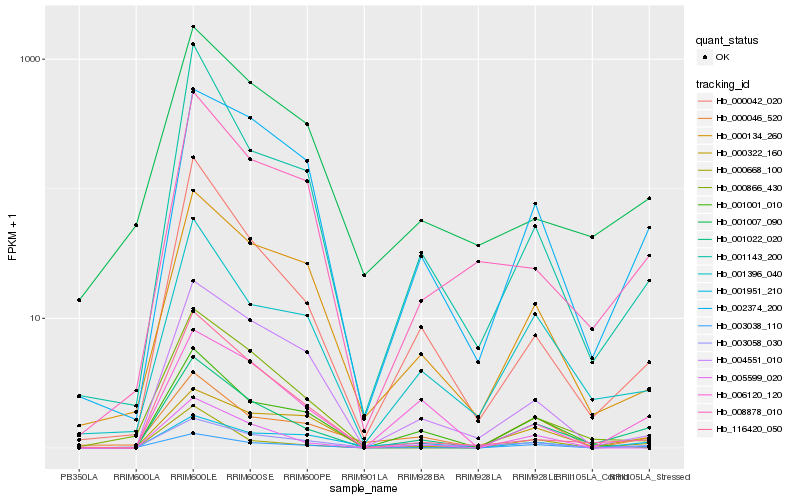
Hb_001022_020 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 2 |
Hb_001396_040 |
0.1665373389 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 3 |
Hb_002374_200 |
0.1666412975 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 4 |
Hb_003038_110 |
0.1681430981 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 5 |
Hb_000042_020 |
0.1758728767 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_006120_120 |
0.1848623631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000668_100 |
0.1945440523 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 8 |
Hb_000134_260 |
0.1948511432 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_008878_010 |
0.1965761568 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 10 |
Hb_005599_020 |
0.2002053918 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 11 |
Hb_003058_030 |
0.2015040906 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001001_010 |
0.202202057 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 13 |
Hb_001951_210 |
0.2056094493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000046_520 |
0.205816479 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004551_010 |
0.2071585081 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 16 |
Hb_000866_430 |
0.2078165337 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 17 |
Hb_001143_200 |
0.2079231301 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 18 |
Hb_001007_090 |
0.2090158592 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 19 |
Hb_000322_160 |
0.2117730249 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2-like [Jatropha curcas] |
| 20 |
Hb_116420_050 |
0.2136195056 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |