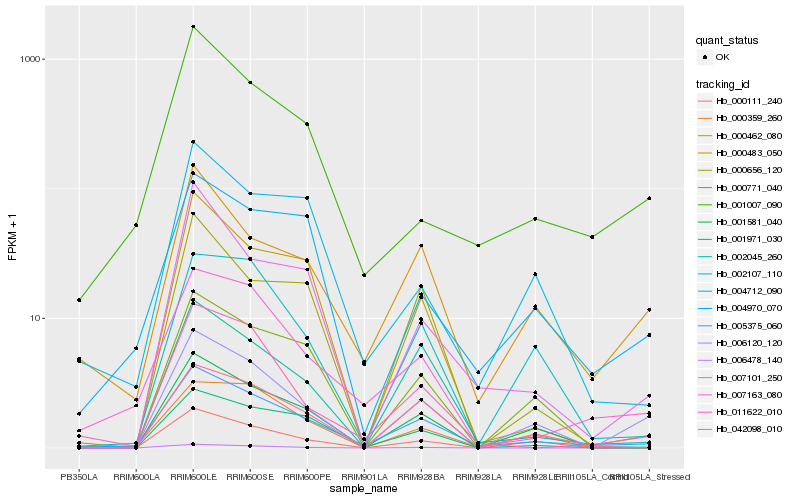
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005375_060 |
0.0 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 2 |
Hb_001581_040 |
0.1413103999 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_007163_080 |
0.1434534998 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 4 |
Hb_002045_260 |
0.151724424 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 5 |
Hb_000462_080 |
0.1523767621 |
- |
- |
PREDICTED: quercetin 3-O-methyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_011622_010 |
0.1583612674 |
- |
- |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 7 |
Hb_042098_010 |
0.160198324 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 8 |
Hb_001971_030 |
0.1645187879 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 9 |
Hb_006120_120 |
0.1686946643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000656_120 |
0.1692044766 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 11 |
Hb_000771_040 |
0.1715967327 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 12 |
Hb_004970_070 |
0.1726636128 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 13 |
Hb_000359_260 |
0.1739552778 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 14 |
Hb_002107_110 |
0.1743646712 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000483_050 |
0.1763329437 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 16 |
Hb_004712_090 |
0.177459181 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 17 |
Hb_007101_250 |
0.1777236596 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 18 |
Hb_000111_240 |
0.1797439682 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 19 |
Hb_001007_090 |
0.182187938 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 20 |
Hb_006478_140 |
0.1842143588 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |