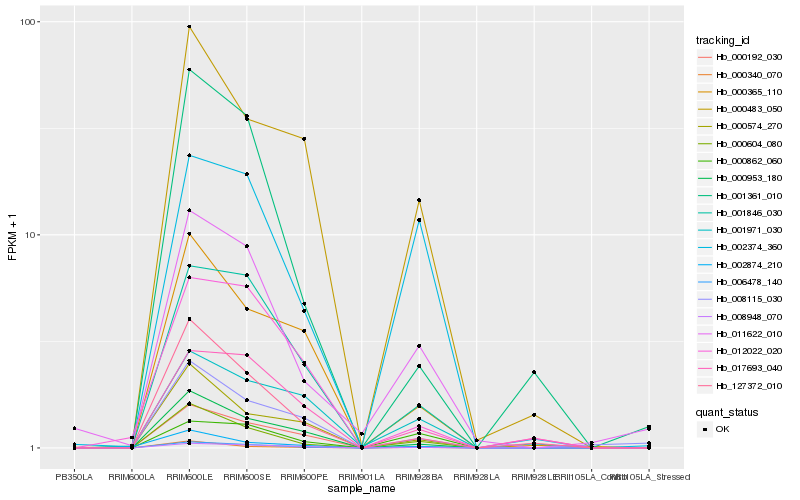
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006478_140 |
0.0 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 2 |
Hb_000604_080 |
0.0870354801 |
- |
- |
Fgenesh protein [Medicago truncatula] |
| 3 |
Hb_008948_070 |
0.1100276707 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001846_030 |
0.1117275208 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 5 |
Hb_000953_180 |
0.1238708957 |
- |
- |
- |
| 6 |
Hb_127372_010 |
0.1407337357 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 7 |
Hb_011622_010 |
0.1440763562 |
- |
- |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 8 |
Hb_000483_050 |
0.1458205381 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 9 |
Hb_000574_270 |
0.1497416565 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 10 |
Hb_000365_110 |
0.1528273669 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 11 |
Hb_000192_030 |
0.1528740003 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 12 |
Hb_017693_040 |
0.1681311246 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 13 |
Hb_001971_030 |
0.1701855865 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 14 |
Hb_008115_030 |
0.1708202189 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001361_010 |
0.172495195 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 16 |
Hb_002374_360 |
0.1726556674 |
- |
- |
hypothetical protein POPTR_0002s08300g [Populus trichocarpa] |
| 17 |
Hb_000862_060 |
0.1757503321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000340_070 |
0.1785157173 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 19 |
Hb_012022_020 |
0.1802545169 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 20 |
Hb_002874_210 |
0.1817927791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |