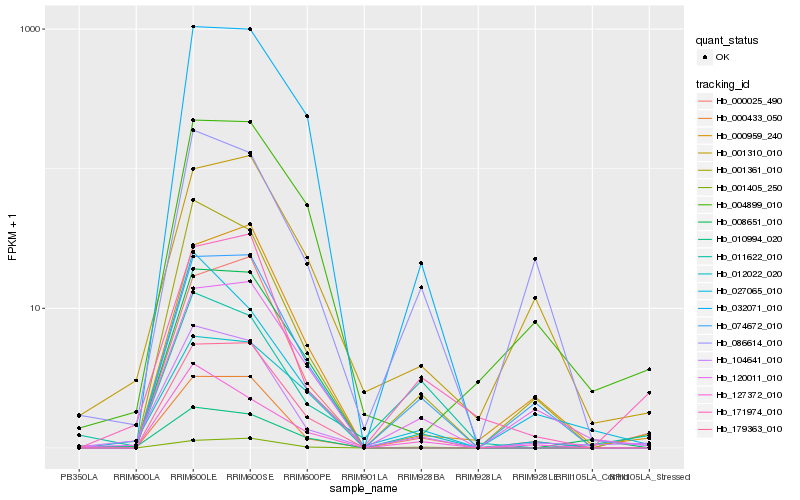
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104641_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001361_010 |
0.1084590191 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 3 |
Hb_074672_010 |
0.1552973733 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 4 |
Hb_171974_010 |
0.1602972304 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 5 |
Hb_000433_050 |
0.1668716375 |
- |
- |
PREDICTED: cell number regulator 1 isoform X1 [Vitis vinifera] |
| 6 |
Hb_120011_010 |
0.1720310733 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_000025_490 |
0.1726734319 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 8 |
Hb_127372_010 |
0.1737131093 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 9 |
Hb_086614_010 |
0.1750825298 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_027065_010 |
0.1770869134 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 11 |
Hb_004899_010 |
0.1813808649 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 12 |
Hb_001405_250 |
0.1826347948 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 13 |
Hb_179363_010 |
0.1829541333 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_008651_010 |
0.183113749 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_012022_020 |
0.1848505622 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 16 |
Hb_011622_010 |
0.1850616421 |
- |
- |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 17 |
Hb_010994_020 |
0.187140518 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000959_240 |
0.1876399071 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_032071_010 |
0.1876833268 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 20 |
Hb_001310_010 |
0.1882058949 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |