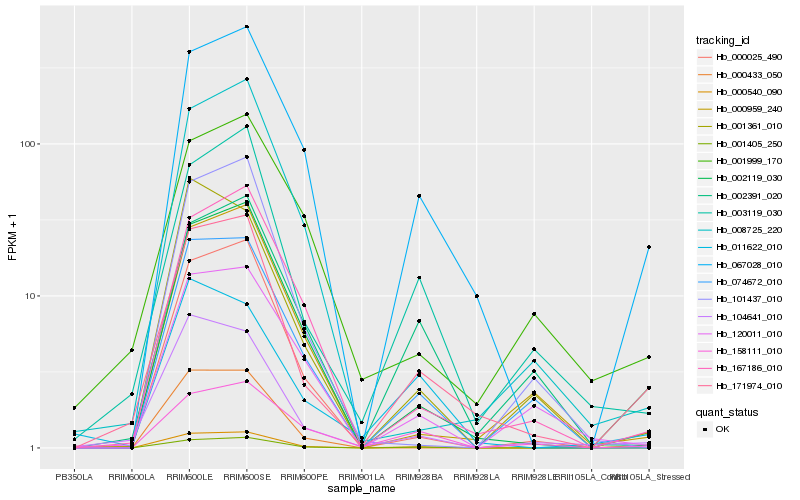
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171974_010 |
0.0 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 2 |
Hb_002119_030 |
0.1306325186 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 3 |
Hb_067028_010 |
0.1340315179 |
- |
- |
BnaC03g72040D [Brassica napus] |
| 4 |
Hb_003119_030 |
0.1450108199 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Jatropha curcas] |
| 5 |
Hb_000540_090 |
0.1498861157 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 6 |
Hb_001405_250 |
0.1534361476 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 7 |
Hb_000025_490 |
0.1571539701 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 8 |
Hb_104641_010 |
0.1602972304 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_167186_010 |
0.1746493715 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 10 |
Hb_074672_010 |
0.1809452523 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_002391_020 |
0.1815636141 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 12 |
Hb_000959_240 |
0.1832770009 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_008725_220 |
0.1893256594 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |
| 14 |
Hb_101437_010 |
0.2007610652 |
- |
- |
PREDICTED: protein P21-like [Jatropha curcas] |
| 15 |
Hb_158111_010 |
0.2012585422 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 16 |
Hb_001999_170 |
0.2036215055 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 17 |
Hb_000433_050 |
0.2040140896 |
- |
- |
PREDICTED: cell number regulator 1 isoform X1 [Vitis vinifera] |
| 18 |
Hb_120011_010 |
0.2044287774 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_001361_010 |
0.2062444372 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 20 |
Hb_011622_010 |
0.2074546651 |
- |
- |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |