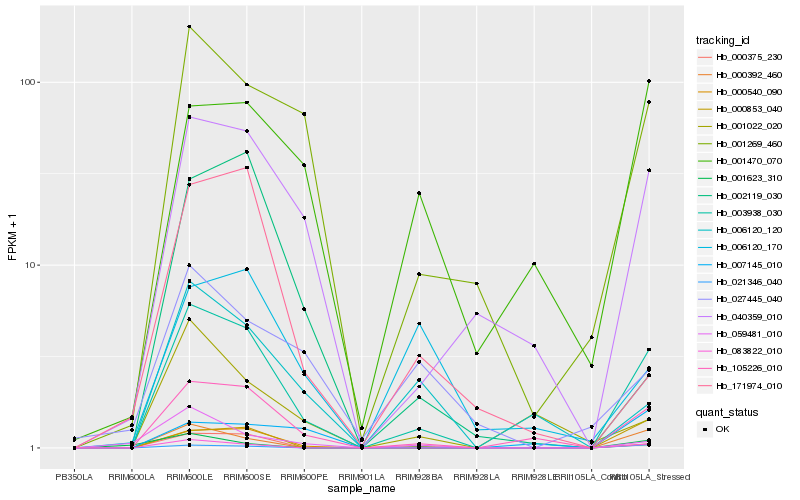
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003938_030 |
0.0 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 2 |
Hb_105226_010 |
0.1511018483 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 3 |
Hb_000540_090 |
0.1897180425 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 4 |
Hb_083822_010 |
0.1928970651 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_040359_010 |
0.19770399 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 6 |
Hb_001269_460 |
0.2124018033 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 7 |
Hb_000375_230 |
0.2159374841 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 8 |
Hb_000392_460 |
0.2355915964 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 9 |
Hb_021346_040 |
0.2433366866 |
- |
- |
hypothetical protein JCGZ_26751 [Jatropha curcas] |
| 10 |
Hb_059481_010 |
0.27839048 |
- |
- |
hypothetical protein POPTR_0008s06860g [Populus trichocarpa] |
| 11 |
Hb_006120_120 |
0.2797900998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000853_040 |
0.2813883737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001470_070 |
0.2821078442 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_027445_040 |
0.3013168339 |
- |
- |
PREDICTED: uncharacterized protein LOC105117996 [Populus euphratica] |
| 15 |
Hb_001022_020 |
0.303919887 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 16 |
Hb_171974_010 |
0.3045174135 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 17 |
Hb_002119_030 |
0.3108175727 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 18 |
Hb_001623_310 |
0.3128234047 |
- |
- |
hypothetical protein CISIN_1g042596mg [Citrus sinensis] |
| 19 |
Hb_007145_010 |
0.3133331216 |
- |
- |
ammonium transporter 3.1-like protein [Camellia sinensis] |
| 20 |
Hb_006120_170 |
0.3158018706 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108-like [Populus euphratica] |