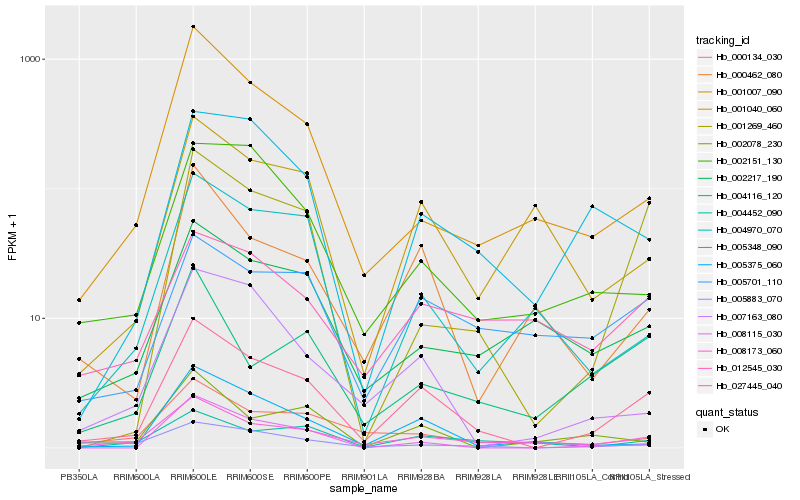
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027445_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105117996 [Populus euphratica] |
| 2 |
Hb_008173_060 |
0.1646314581 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 3 |
Hb_005375_060 |
0.200374314 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 4 |
Hb_000462_080 |
0.2026724306 |
- |
- |
PREDICTED: quercetin 3-O-methyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_007163_080 |
0.213402801 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 6 |
Hb_005348_090 |
0.2140567006 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 7 |
Hb_004452_090 |
0.2141462507 |
- |
- |
hypothetical protein JCGZ_12231 [Jatropha curcas] |
| 8 |
Hb_001269_460 |
0.2145364446 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 9 |
Hb_005701_110 |
0.2196299219 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 10 |
Hb_005883_070 |
0.2198223629 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |
| 11 |
Hb_000134_030 |
0.2205113222 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 12 |
Hb_001007_090 |
0.2213826631 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 13 |
Hb_002151_130 |
0.2236553843 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 14 |
Hb_004970_070 |
0.2255809827 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 15 |
Hb_002217_190 |
0.2265203016 |
- |
- |
invertase [Hevea brasiliensis] |
| 16 |
Hb_008115_030 |
0.2305340198 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001040_060 |
0.2316603157 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 18 |
Hb_004116_120 |
0.2340206399 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 19 |
Hb_012545_030 |
0.2384249807 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 20 |
Hb_002078_230 |
0.2395046069 |
- |
- |
PREDICTED: HORMA domain-containing protein 1 [Jatropha curcas] |