| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
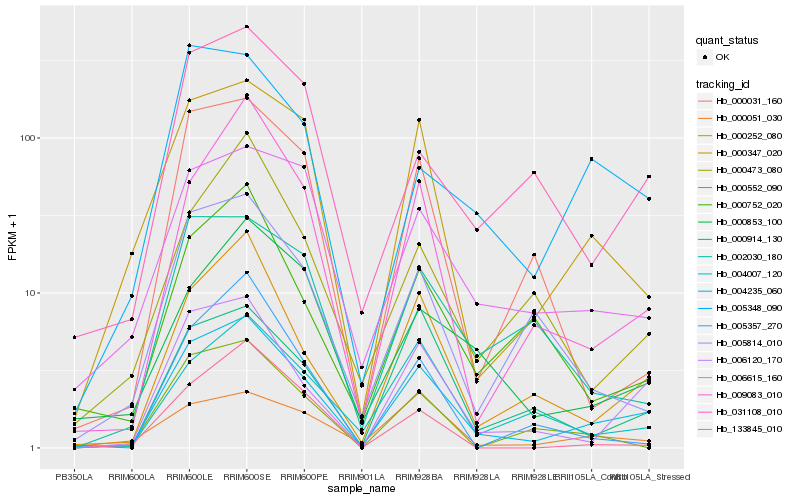
Hb_004235_060 |
0.0 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 2 |
Hb_006120_170 |
0.1341164116 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108-like [Populus euphratica] |
| 3 |
Hb_000252_080 |
0.1489852572 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 4 |
Hb_000347_020 |
0.162658067 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 5 |
Hb_009083_010 |
0.1766443 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 6 |
Hb_006615_160 |
0.1870490224 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 7 |
Hb_031108_010 |
0.188166339 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 8 |
Hb_004007_120 |
0.1899931434 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 9 |
Hb_000914_130 |
0.1919823249 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 10 |
Hb_005348_090 |
0.1921910739 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 11 |
Hb_000752_020 |
0.1947329315 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005814_010 |
0.1961300624 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000031_160 |
0.1982721868 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 14 |
Hb_002030_180 |
0.1992808449 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 15 |
Hb_000853_100 |
0.1992920473 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 16 |
Hb_000552_090 |
0.2000919045 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 17 |
Hb_000051_030 |
0.2011564941 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 18 |
Hb_000473_080 |
0.2071774678 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 19 |
Hb_005357_270 |
0.2087875623 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_133845_010 |
0.2113155866 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |