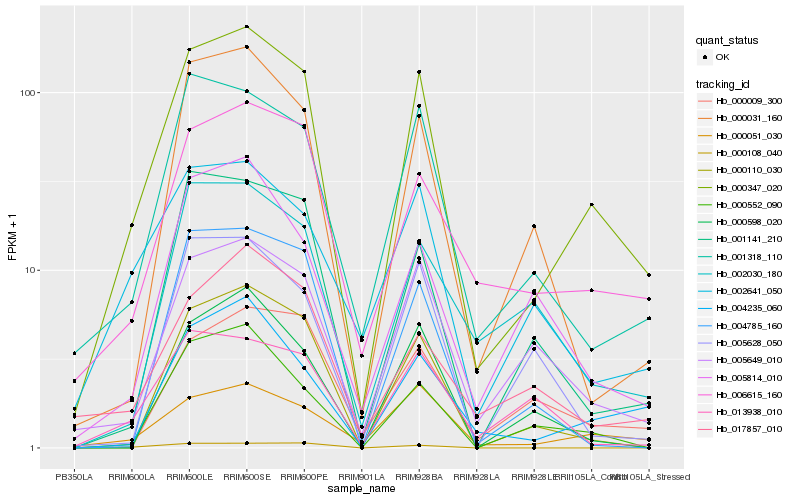
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_020 |
0.0 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 2 |
Hb_000051_030 |
0.1426293502 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 3 |
Hb_001318_110 |
0.1517682881 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 4 |
Hb_006615_160 |
0.153927904 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 5 |
Hb_005649_010 |
0.1617551315 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 6 |
Hb_004235_060 |
0.162658067 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_000110_030 |
0.1716375724 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001141_210 |
0.17737677 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 9 |
Hb_000552_090 |
0.177427645 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 10 |
Hb_000031_160 |
0.1777032051 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 11 |
Hb_002641_050 |
0.177856252 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 12 |
Hb_005814_010 |
0.1898243699 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_017857_010 |
0.1898669787 |
- |
- |
- |
| 14 |
Hb_004785_160 |
0.193559877 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 15 |
Hb_000009_300 |
0.194441811 |
- |
- |
hypothetical protein CICLE_v10024731mg [Citrus clementina] |
| 16 |
Hb_000598_020 |
0.1949342371 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 17 |
Hb_002030_180 |
0.1966060391 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_013938_010 |
0.2014238904 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 19 |
Hb_005628_050 |
0.2015064602 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_000108_040 |
0.2030538466 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Gossypium raimondii] |