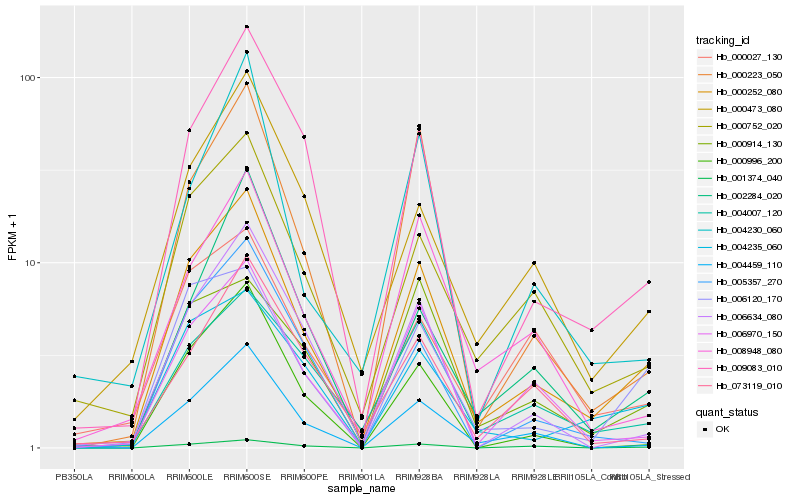
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 2 |
Hb_000752_020 |
0.1316510045 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009083_010 |
0.1361383685 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 4 |
Hb_004459_110 |
0.1401154035 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 5 |
Hb_006970_150 |
0.1422225273 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 6 |
Hb_006120_170 |
0.1467138758 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108-like [Populus euphratica] |
| 7 |
Hb_004235_060 |
0.1489852572 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_000223_050 |
0.1517714639 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 9 |
Hb_006634_080 |
0.154588945 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 10 |
Hb_000473_080 |
0.1546384109 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 11 |
Hb_004007_120 |
0.1585020694 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 12 |
Hb_008948_080 |
0.1590340882 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000996_200 |
0.1639427943 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Populus euphratica] |
| 14 |
Hb_001374_040 |
0.1689240605 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 15 |
Hb_005357_270 |
0.1711345462 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000914_130 |
0.1756481609 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 17 |
Hb_004230_060 |
0.1781152576 |
- |
- |
PREDICTED: ferritin, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_073119_010 |
0.1792376567 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 19 |
Hb_002284_020 |
0.180394807 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 20 |
Hb_000027_130 |
0.1808865828 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |