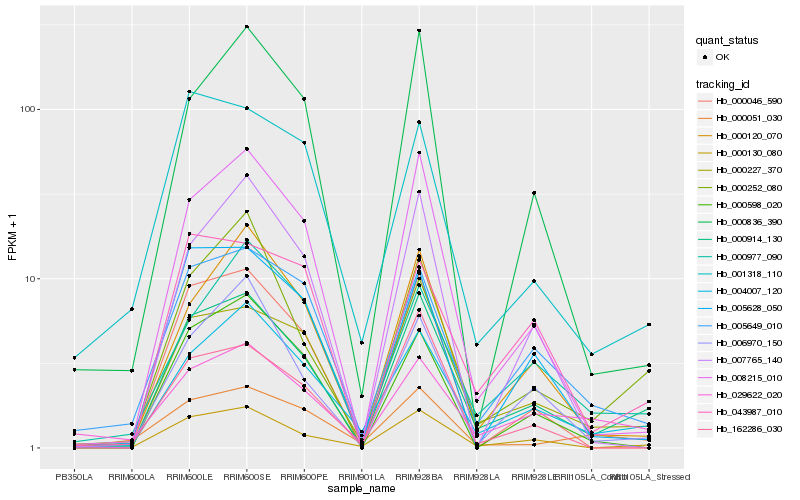
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000914_130 |
0.0 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 2 |
Hb_000130_080 |
0.1196528164 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_004007_120 |
0.1268918027 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 4 |
Hb_008215_010 |
0.137561089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005649_010 |
0.1541649234 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 6 |
Hb_000227_370 |
0.1554895957 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_000120_070 |
0.158890152 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 8 |
Hb_000836_390 |
0.1630730294 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 9 |
Hb_000046_590 |
0.1650459932 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 10 |
Hb_006970_150 |
0.1677119158 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 11 |
Hb_029622_020 |
0.171489911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000252_080 |
0.1756481609 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 13 |
Hb_005628_050 |
0.1766906159 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_043987_010 |
0.1791999262 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 15 |
Hb_000977_090 |
0.1796987388 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 16 |
Hb_001318_110 |
0.1802573192 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 17 |
Hb_162286_030 |
0.1808374697 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 18 |
Hb_000051_030 |
0.1818046183 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 19 |
Hb_000598_020 |
0.1825082639 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 20 |
Hb_007765_140 |
0.1825396054 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |