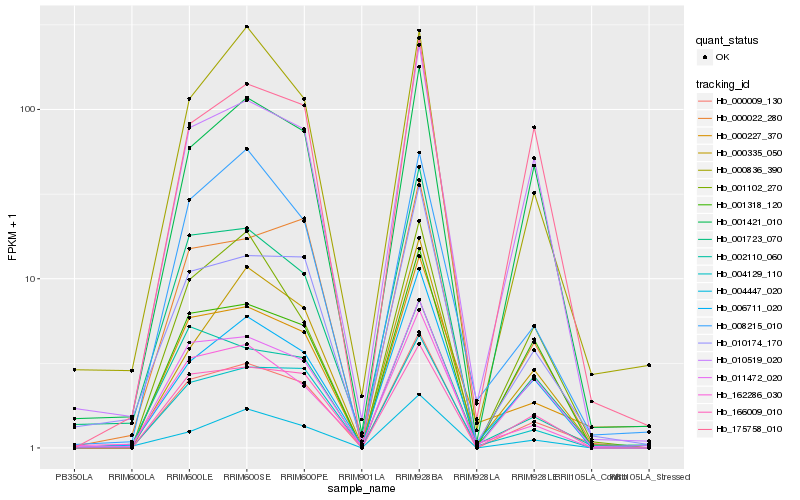
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 2 |
Hb_166009_010 |
0.0954772096 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 3 |
Hb_004129_110 |
0.1037506022 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 4 |
Hb_011472_020 |
0.1084490616 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 5 |
Hb_010519_020 |
0.1127269047 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 6 |
Hb_001421_010 |
0.1156795855 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000022_280 |
0.1174446999 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 8 |
Hb_010174_170 |
0.1180513588 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001318_120 |
0.118837204 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 10 |
Hb_002110_060 |
0.1233611667 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 11 |
Hb_162286_030 |
0.1254751726 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 12 |
Hb_000227_370 |
0.1259858835 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 13 |
Hb_004447_020 |
0.1281755342 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 14 |
Hb_175758_010 |
0.128950542 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000335_050 |
0.1312340403 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001102_270 |
0.133107775 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_006711_020 |
0.1351361494 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 18 |
Hb_001723_070 |
0.1369955581 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 19 |
Hb_000836_390 |
0.1384859855 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 20 |
Hb_008215_010 |
0.1408765887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |