| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
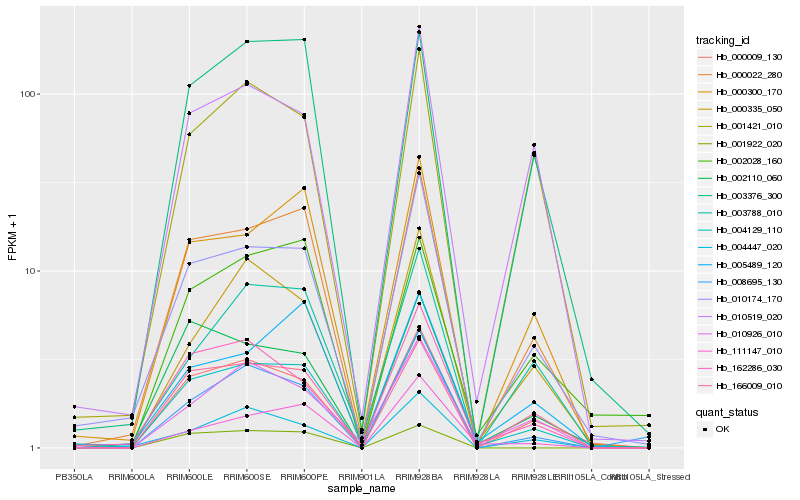
Hb_004129_110 |
0.0 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 2 |
Hb_000022_280 |
0.0619671071 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 3 |
Hb_166009_010 |
0.0973528962 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 4 |
Hb_000009_130 |
0.1037506022 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 5 |
Hb_010174_170 |
0.1085282942 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000300_170 |
0.1108179104 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 7 |
Hb_003376_300 |
0.1219639075 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 8 |
Hb_002110_060 |
0.1292069391 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 9 |
Hb_162286_030 |
0.1340882112 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 10 |
Hb_010519_020 |
0.1382333198 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 11 |
Hb_004447_020 |
0.140412349 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 12 |
Hb_001421_010 |
0.1425016701 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010926_010 |
0.1435286706 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 14 |
Hb_000335_050 |
0.1459137058 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008695_130 |
0.1471073387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003788_010 |
0.1472884182 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 17 |
Hb_111147_010 |
0.1479620595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005489_120 |
0.1483493817 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 19 |
Hb_001922_020 |
0.1484317117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002028_160 |
0.1493424071 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Populus euphratica] |