| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
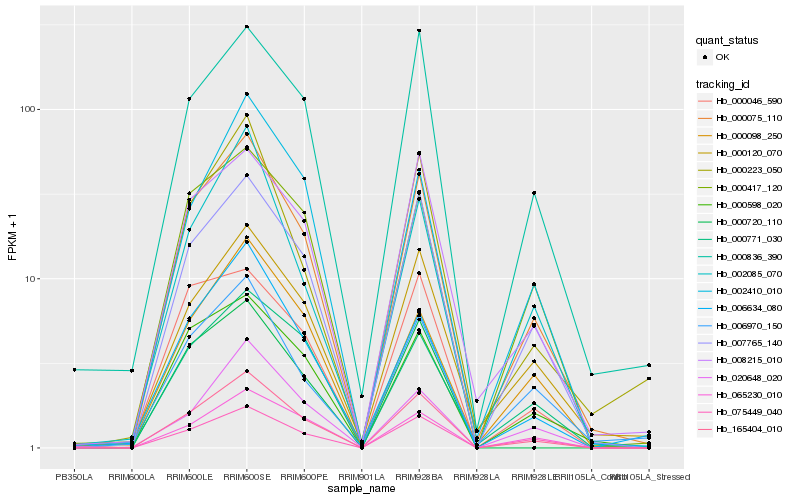
| 1 |
Hb_165404_010 |
0.0 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 2 |
Hb_000075_110 |
0.0484504823 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 3 |
Hb_007765_140 |
0.0724784696 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 4 |
Hb_065230_010 |
0.1049451129 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 5 |
Hb_008215_010 |
0.1091300415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_075449_040 |
0.1117744247 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 7 |
Hb_000120_070 |
0.1123064531 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 8 |
Hb_000598_020 |
0.1126577921 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 9 |
Hb_000836_390 |
0.1153771898 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 10 |
Hb_000771_030 |
0.1209248984 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000720_110 |
0.1222631805 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 12 |
Hb_000417_120 |
0.1226315671 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 13 |
Hb_002410_010 |
0.1226673379 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 14 |
Hb_006634_080 |
0.1235275674 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 15 |
Hb_000223_050 |
0.1281138597 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 16 |
Hb_020648_020 |
0.1328547503 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 17 |
Hb_002085_070 |
0.1346126718 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000098_250 |
0.1349839287 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 19 |
Hb_000046_590 |
0.1362801293 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_006970_150 |
0.1372400706 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |