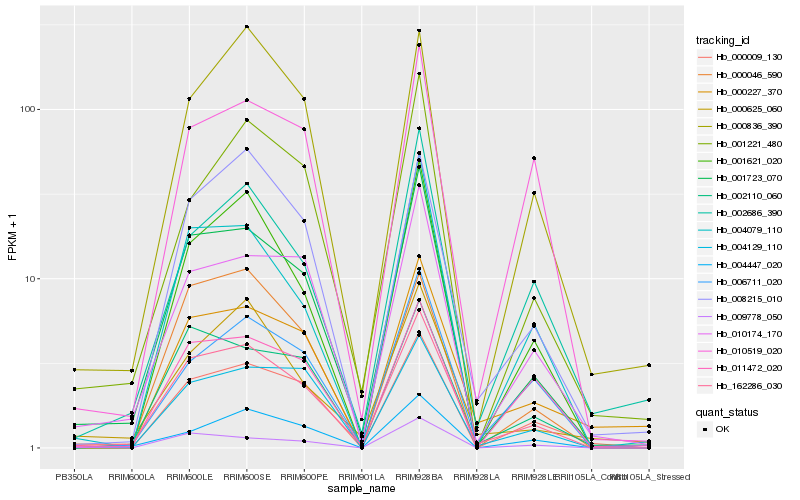
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162286_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 2 |
Hb_001723_070 |
0.0731115794 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 3 |
Hb_001621_020 |
0.1186698206 |
- |
- |
- |
| 4 |
Hb_002110_060 |
0.1221554902 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 5 |
Hb_000009_130 |
0.1254751726 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 6 |
Hb_010174_170 |
0.129565444 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008215_010 |
0.129834962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004079_110 |
0.1329604677 |
- |
- |
PREDICTED: uncharacterized protein LOC100257802 [Vitis vinifera] |
| 9 |
Hb_004129_110 |
0.1340882112 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 10 |
Hb_009778_050 |
0.1363708917 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 11 |
Hb_006711_020 |
0.1389694757 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 12 |
Hb_000046_590 |
0.1399633369 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 13 |
Hb_010519_020 |
0.1407546785 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 14 |
Hb_002686_390 |
0.1411443885 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 15 |
Hb_004447_020 |
0.1414480073 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 16 |
Hb_000227_370 |
0.1440239774 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_000836_390 |
0.1456308329 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 18 |
Hb_011472_020 |
0.1462087799 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 19 |
Hb_000625_060 |
0.1466273135 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 20 |
Hb_001221_480 |
0.1468649701 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |