| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
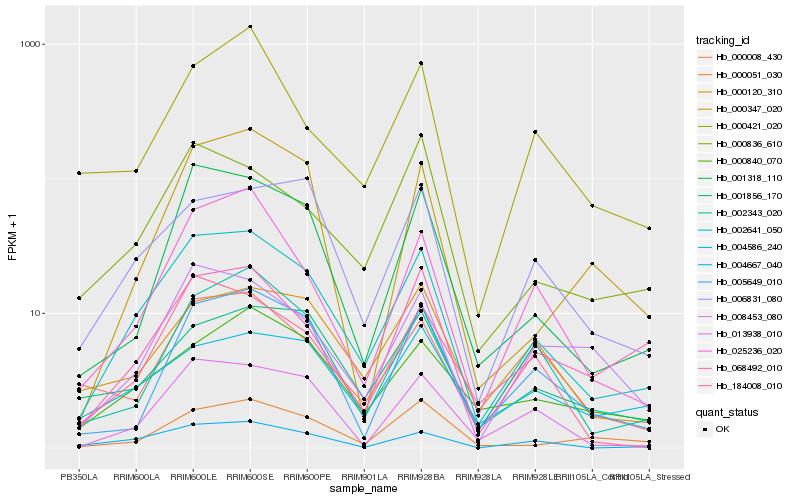
Hb_002641_050 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 2 |
Hb_001318_110 |
0.1372406527 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 3 |
Hb_004667_040 |
0.1413156757 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_025236_020 |
0.1454499447 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 5 |
Hb_013938_010 |
0.1474562887 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 6 |
Hb_002343_020 |
0.1530377383 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 7 |
Hb_184008_010 |
0.155324341 |
- |
- |
putative LRR receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 8 |
Hb_008453_080 |
0.1588924273 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X2 [Jatropha curcas] |
| 9 |
Hb_006831_080 |
0.160196882 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 10 |
Hb_000840_070 |
0.163654044 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005649_010 |
0.1644329428 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 12 |
Hb_000836_610 |
0.1647041344 |
- |
- |
stem-specific protein TSJT1-like [Jatropha curcas] |
| 13 |
Hb_068492_010 |
0.1654513274 |
- |
- |
- |
| 14 |
Hb_001856_170 |
0.1665948778 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 15 |
Hb_000120_310 |
0.1683792295 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 16 |
Hb_000051_030 |
0.171030022 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 17 |
Hb_004586_240 |
0.1741280164 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 18 |
Hb_000421_020 |
0.1770285458 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 19 |
Hb_000008_430 |
0.1775138194 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000347_020 |
0.177856252 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |