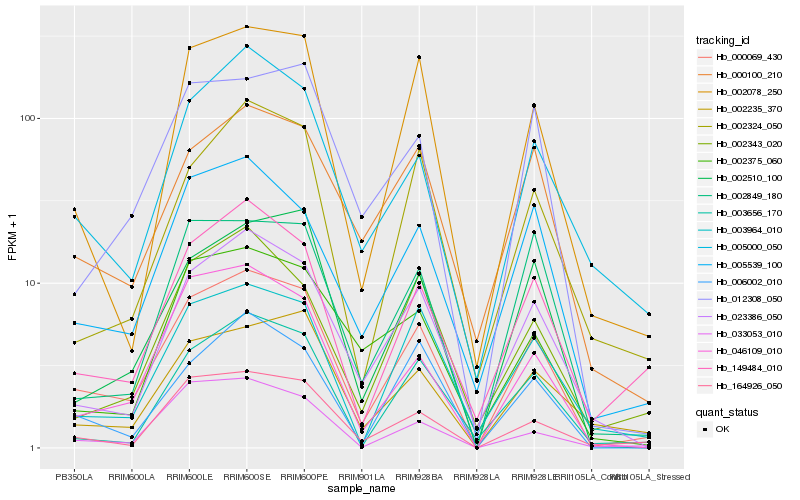
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_430 |
0.0 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 2 |
Hb_164926_050 |
0.1254516736 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 3 |
Hb_002078_250 |
0.1260270763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_149484_010 |
0.1264295667 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 5 |
Hb_005539_100 |
0.1288379387 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 6 |
Hb_023386_050 |
0.1304225861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005000_050 |
0.1327151148 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 8 |
Hb_046109_010 |
0.1334687015 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_002510_100 |
0.1347721392 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 10 |
Hb_003964_010 |
0.1363215114 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 11 |
Hb_000100_210 |
0.1384413842 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 12 |
Hb_006002_010 |
0.1421191201 |
transcription factor |
TF Family: MYB |
Myb domain protein 5 isoform 1 [Theobroma cacao] |
| 13 |
Hb_002235_370 |
0.1439129148 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 14 |
Hb_012308_050 |
0.1448967038 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 15 |
Hb_003656_170 |
0.1466131552 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 16 |
Hb_002324_050 |
0.1483520999 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 17 |
Hb_002849_180 |
0.1493343356 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 18 |
Hb_002343_020 |
0.1501047218 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 19 |
Hb_033053_010 |
0.1542917903 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 20 |
Hb_002375_060 |
0.1559652382 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |