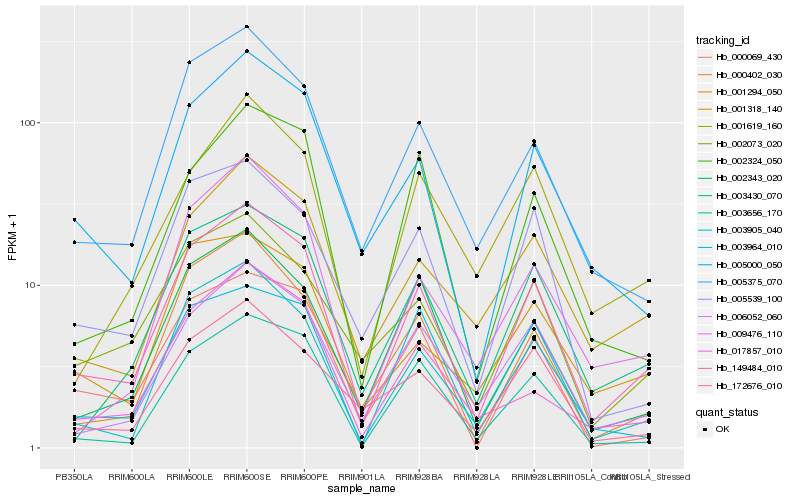
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_149484_010 |
0.0 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 2 |
Hb_003905_040 |
0.1017340964 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 3 |
Hb_006052_060 |
0.1119975325 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 4 |
Hb_003430_070 |
0.1123990468 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 5 |
Hb_005000_050 |
0.1143695137 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 6 |
Hb_000402_030 |
0.1145635238 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 7 |
Hb_005375_070 |
0.1166282945 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 8 |
Hb_001318_140 |
0.1191423795 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 9 |
Hb_002343_020 |
0.1223052947 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 10 |
Hb_003656_170 |
0.1246592019 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 11 |
Hb_000069_430 |
0.1264295667 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 12 |
Hb_002073_020 |
0.1287135212 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 13 |
Hb_005539_100 |
0.1309580672 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 14 |
Hb_009476_110 |
0.1311383404 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 15 |
Hb_172676_010 |
0.132706539 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_002324_050 |
0.1350120847 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 17 |
Hb_001619_160 |
0.1356315797 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 18 |
Hb_001294_050 |
0.1363473543 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 19 |
Hb_017857_010 |
0.1377772016 |
- |
- |
- |
| 20 |
Hb_003964_010 |
0.1420486487 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |