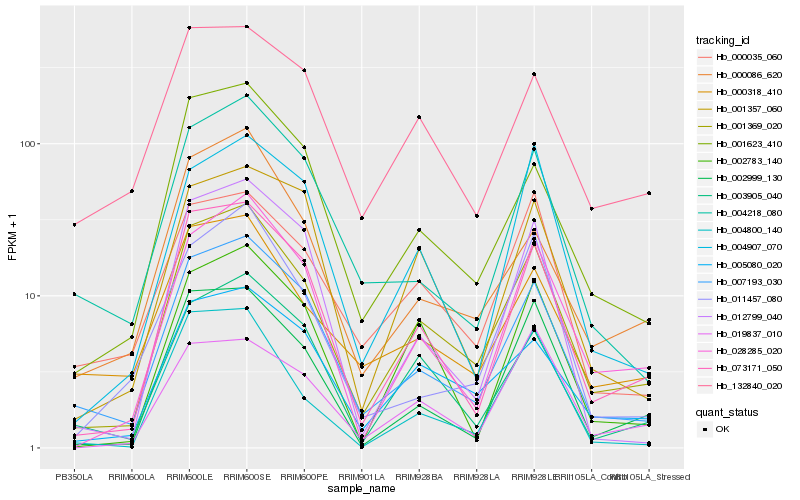
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_020 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 2 |
Hb_073171_050 |
0.0857344592 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 3 |
Hb_007193_030 |
0.0943953065 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 4 |
Hb_028285_020 |
0.1034334809 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_005080_020 |
0.1075236293 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 6 |
Hb_001623_410 |
0.1160494797 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003905_040 |
0.120773943 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 8 |
Hb_002999_130 |
0.1266650821 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 9 |
Hb_002783_140 |
0.1326268725 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 10 |
Hb_000318_410 |
0.1381092907 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 11 |
Hb_004907_070 |
0.1415206375 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 12 |
Hb_000086_620 |
0.1427592024 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_132840_020 |
0.1430721274 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 14 |
Hb_000035_060 |
0.1450168175 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001357_060 |
0.1452203419 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 16 |
Hb_004218_080 |
0.1464253585 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |
| 17 |
Hb_012799_040 |
0.1494199348 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 18 |
Hb_004800_140 |
0.149534755 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011457_080 |
0.1509415191 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_019837_010 |
0.1515229328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |