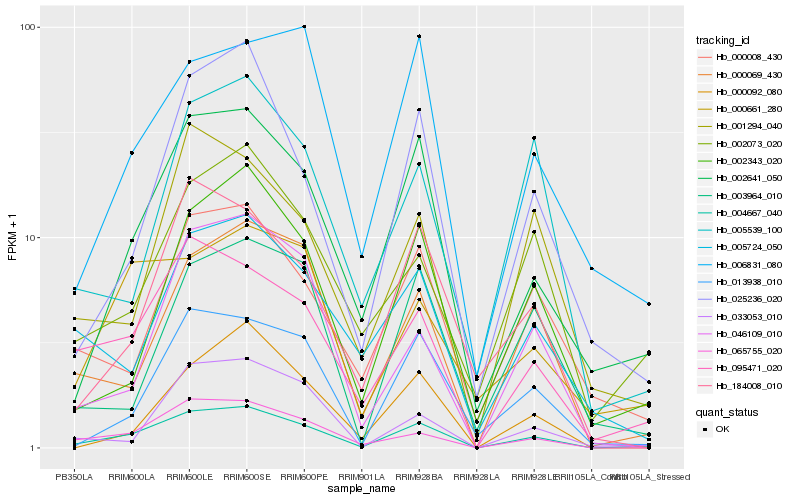
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004667_040 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_002641_050 |
0.1413156757 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 3 |
Hb_095471_020 |
0.1468015424 |
- |
- |
- |
| 4 |
Hb_065755_020 |
0.1511049918 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 5 |
Hb_000069_430 |
0.1658290126 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 6 |
Hb_025236_020 |
0.169358766 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 7 |
Hb_002343_020 |
0.1695532987 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 8 |
Hb_013938_010 |
0.171534991 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 9 |
Hb_000008_430 |
0.1838538643 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 10 |
Hb_184008_010 |
0.185837991 |
- |
- |
putative LRR receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 11 |
Hb_046109_010 |
0.1864110815 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_002073_020 |
0.1867499044 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 13 |
Hb_001294_040 |
0.1888715788 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 14 |
Hb_033053_010 |
0.190809021 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 15 |
Hb_005724_050 |
0.1955587561 |
- |
- |
- |
| 16 |
Hb_003964_010 |
0.1955765407 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 17 |
Hb_006831_080 |
0.1957311849 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 18 |
Hb_000661_280 |
0.1958944873 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 19 |
Hb_000092_080 |
0.1961204654 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_038742 [Vitis vinifera] |
| 20 |
Hb_005539_100 |
0.1970448776 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |