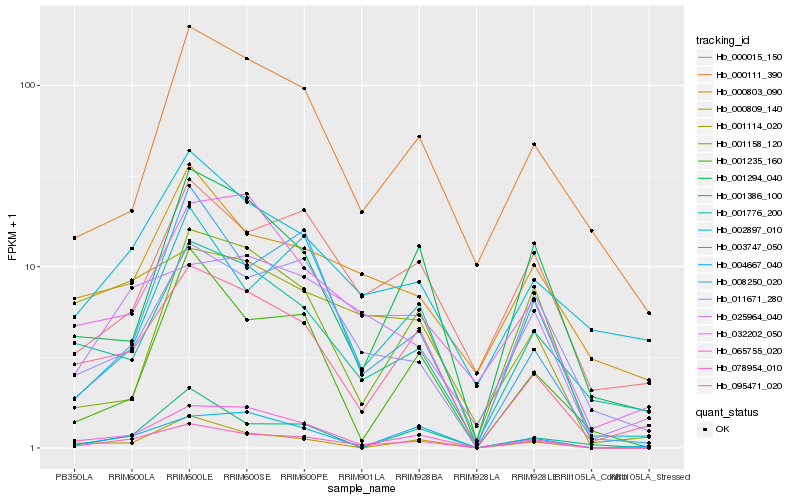
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078954_010 |
0.0 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 2 |
Hb_065755_020 |
0.146038979 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_095471_020 |
0.1573904007 |
- |
- |
- |
| 4 |
Hb_002897_010 |
0.1766008876 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 5 |
Hb_011671_280 |
0.1905906831 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF106-like [Populus euphratica] |
| 6 |
Hb_000809_140 |
0.1908164823 |
- |
- |
hypothetical protein JCGZ_12354 [Jatropha curcas] |
| 7 |
Hb_003747_050 |
0.1941977532 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 8 |
Hb_001294_040 |
0.1945802468 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 9 |
Hb_008250_020 |
0.195889368 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000015_150 |
0.1971038696 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 11 |
Hb_004667_040 |
0.1979569189 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_001776_200 |
0.1986845975 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 13 |
Hb_025964_040 |
0.2002791212 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 14 |
Hb_001235_160 |
0.2032660929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001114_020 |
0.2050747021 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 16 |
Hb_001386_100 |
0.2060506359 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor PHERES 1-like [Jatropha curcas] |
| 17 |
Hb_000111_390 |
0.2074706111 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001158_120 |
0.2090000718 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 19 |
Hb_032202_050 |
0.2097242557 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000803_090 |
0.2119567952 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |