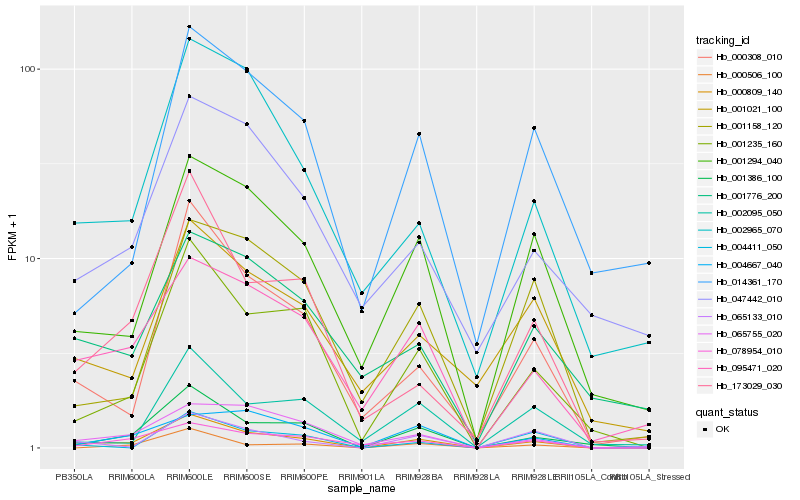
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_12354 [Jatropha curcas] |
| 2 |
Hb_001386_100 |
0.1496909737 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor PHERES 1-like [Jatropha curcas] |
| 3 |
Hb_001235_160 |
0.1545180869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_095471_020 |
0.1751193356 |
- |
- |
- |
| 5 |
Hb_000308_010 |
0.1770028473 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 6 |
Hb_065755_020 |
0.1841947989 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_001294_040 |
0.1865813867 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 8 |
Hb_173029_030 |
0.1901469976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_078954_010 |
0.1908164823 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 10 |
Hb_002965_070 |
0.1916380411 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 11 |
Hb_065133_010 |
0.2078447981 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 12 |
Hb_001021_100 |
0.2118746819 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 13 |
Hb_000506_100 |
0.2181306572 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g48130 [Jatropha curcas] |
| 14 |
Hb_001776_200 |
0.2199922267 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 15 |
Hb_047442_010 |
0.2206997788 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 16 |
Hb_002095_050 |
0.2212999431 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 17 |
Hb_001158_120 |
0.2218194805 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004667_040 |
0.2237748236 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_004411_050 |
0.2260468093 |
- |
- |
quinone oxidoreductase-like protein [Arabidopsis thaliana] |
| 20 |
Hb_014361_170 |
0.2262686296 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |