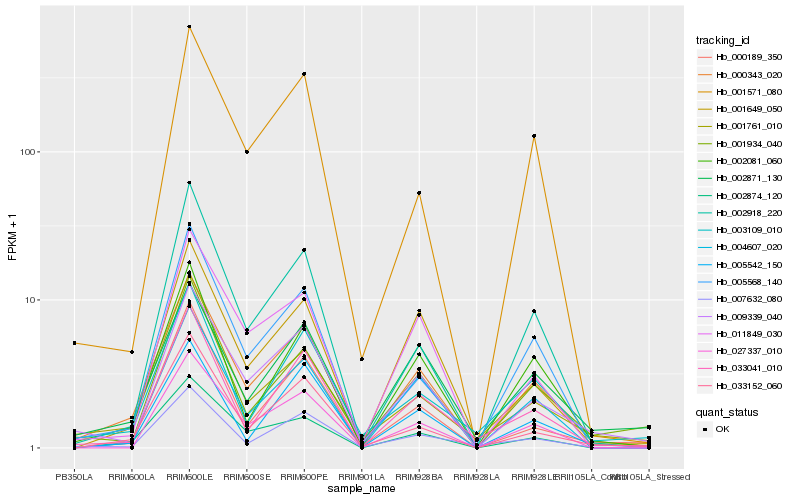
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_020 |
0.0 |
- |
- |
PREDICTED: putative Peroxidase 48 [Jatropha curcas] |
| 2 |
Hb_002874_120 |
0.108308628 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 3 |
Hb_009339_040 |
0.1128789029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002081_060 |
0.1187164791 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 5 |
Hb_004607_020 |
0.1196952542 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 6 |
Hb_011849_030 |
0.1308377468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_033152_060 |
0.1313344838 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 8 |
Hb_005568_140 |
0.131747962 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_033041_010 |
0.1329414496 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 10 |
Hb_027337_010 |
0.1334485139 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 11 |
Hb_002918_220 |
0.1340105796 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 12 |
Hb_001649_050 |
0.1363170974 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 13 |
Hb_002871_130 |
0.1392971503 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 14 |
Hb_001761_010 |
0.1407045871 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 15 |
Hb_001934_040 |
0.1425283727 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like [Jatropha curcas] |
| 16 |
Hb_005542_150 |
0.1427672877 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 17 |
Hb_001571_080 |
0.1430600942 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 18 |
Hb_007632_080 |
0.144391412 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 19 |
Hb_003109_010 |
0.1496701529 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 20 |
Hb_000189_350 |
0.1499262966 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |