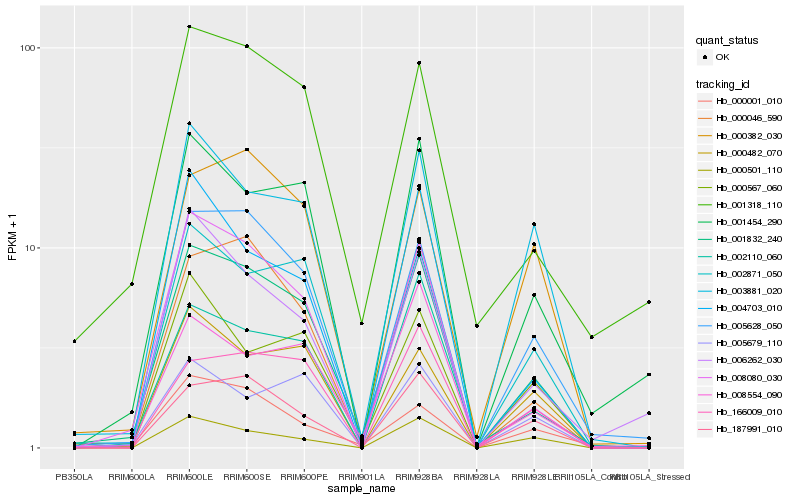
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001832_240 |
0.0 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 2 |
Hb_008080_030 |
0.104548833 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 3 |
Hb_005628_050 |
0.1049719822 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_002871_050 |
0.1064182971 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 5 |
Hb_000046_590 |
0.114696405 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_001454_290 |
0.1357900669 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 7 |
Hb_003881_020 |
0.1366794457 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 8 |
Hb_002110_060 |
0.1375026107 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 9 |
Hb_187991_010 |
0.1401910101 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 10 |
Hb_001318_110 |
0.1461256083 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 11 |
Hb_000482_070 |
0.148309851 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 12 |
Hb_000001_010 |
0.1491664272 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 13 |
Hb_004703_010 |
0.1510678902 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 14 |
Hb_005679_110 |
0.1511908795 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 15 |
Hb_166009_010 |
0.1512415202 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 16 |
Hb_000567_060 |
0.153531748 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 17 |
Hb_000382_030 |
0.1556063517 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 18 |
Hb_006262_030 |
0.1579902499 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_008554_090 |
0.1582387887 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000501_110 |
0.1593776247 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |