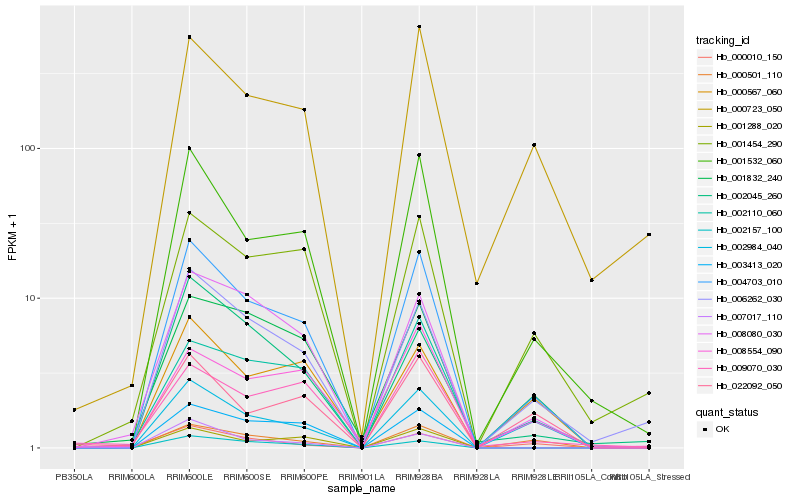
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004703_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 2 |
Hb_001532_060 |
0.0895790521 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_006262_030 |
0.1284293299 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_008080_030 |
0.1287851434 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 5 |
Hb_003413_020 |
0.1435939498 |
- |
- |
- |
| 6 |
Hb_002110_060 |
0.1456799061 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 7 |
Hb_002157_100 |
0.1465028332 |
- |
- |
- |
| 8 |
Hb_001288_020 |
0.1490720633 |
- |
- |
- |
| 9 |
Hb_001832_240 |
0.1510678902 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 10 |
Hb_000501_110 |
0.1515566479 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 11 |
Hb_001454_290 |
0.1540475053 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 12 |
Hb_002984_040 |
0.1549305893 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_007017_110 |
0.159215414 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 14 |
Hb_000567_060 |
0.1595526934 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 15 |
Hb_000723_050 |
0.1621942152 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 16 |
Hb_022092_050 |
0.1660213084 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 17 |
Hb_002045_260 |
0.1673130164 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 18 |
Hb_008554_090 |
0.1692584844 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000010_150 |
0.172451983 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Jatropha curcas] |
| 20 |
Hb_009070_030 |
0.1734430649 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |