| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
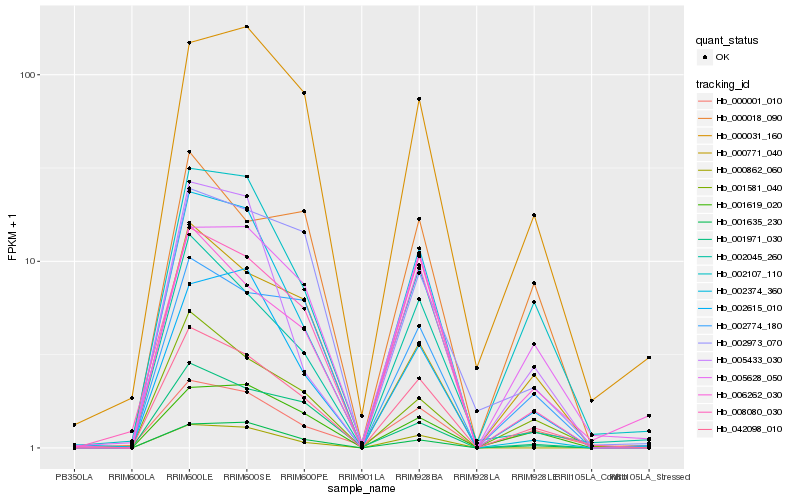
Hb_042098_010 |
0.0 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 2 |
Hb_001581_040 |
0.1101017937 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002045_260 |
0.1244852084 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 4 |
Hb_008080_030 |
0.1259708022 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 5 |
Hb_005628_050 |
0.1290761746 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_000001_010 |
0.1301285731 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 7 |
Hb_002774_180 |
0.1325049102 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_001635_230 |
0.1326016693 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002107_110 |
0.1347364939 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006262_030 |
0.1359064381 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000771_040 |
0.1387582876 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 12 |
Hb_001619_020 |
0.1450091417 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 13 |
Hb_002615_010 |
0.1480610992 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 14 |
Hb_002374_360 |
0.1534016297 |
- |
- |
hypothetical protein POPTR_0002s08300g [Populus trichocarpa] |
| 15 |
Hb_000031_160 |
0.1542541639 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 16 |
Hb_000018_090 |
0.1573534073 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000862_060 |
0.1575986688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001971_030 |
0.1578004565 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 19 |
Hb_002973_070 |
0.1584298404 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 20 |
Hb_005433_030 |
0.1593254272 |
transcription factor |
TF Family: MYB |
unnamed protein product [Vitis vinifera] |