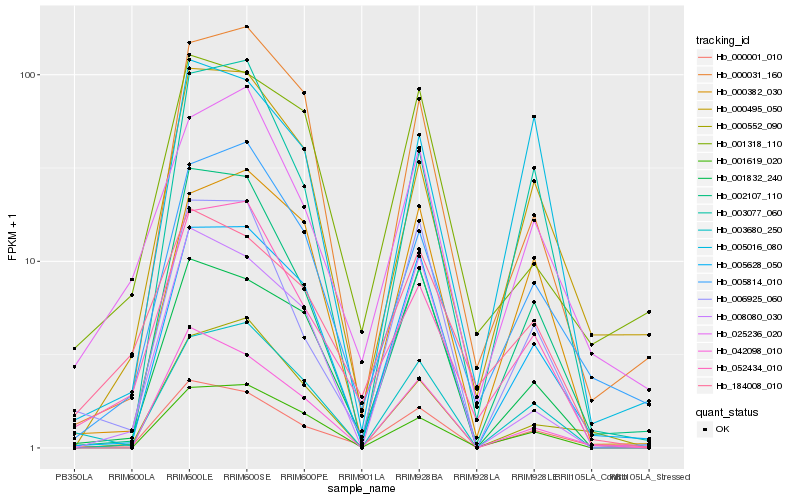
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 2 |
Hb_005628_050 |
0.121874699 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_006925_060 |
0.1299283531 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 4 |
Hb_042098_010 |
0.1301285731 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 5 |
Hb_052434_010 |
0.1367194157 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 6 |
Hb_002107_110 |
0.1408915842 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000495_050 |
0.1476655175 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 8 |
Hb_001832_240 |
0.1491664272 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 9 |
Hb_025236_020 |
0.1503892834 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 10 |
Hb_005814_010 |
0.1511383324 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_184008_010 |
0.1594857058 |
- |
- |
putative LRR receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 12 |
Hb_003680_250 |
0.1596629804 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_003077_060 |
0.160717332 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 14 |
Hb_001619_020 |
0.1615470234 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 15 |
Hb_000552_090 |
0.1667784856 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 16 |
Hb_005016_080 |
0.1694535007 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 17 |
Hb_001318_110 |
0.1704900055 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 18 |
Hb_000382_030 |
0.1722988765 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 19 |
Hb_008080_030 |
0.1731370858 |
- |
- |
hypothetical protein POPTR_0275s002101g, partial [Populus trichocarpa] |
| 20 |
Hb_000031_160 |
0.1733935244 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |