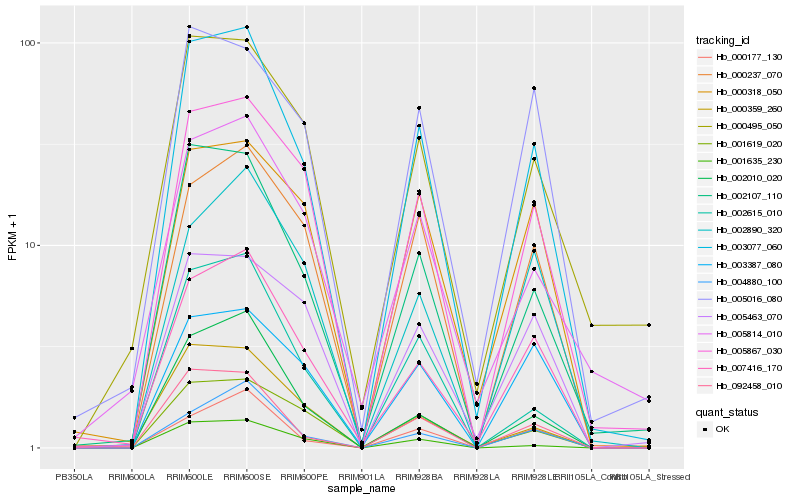
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003077_060 |
0.0 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 2 |
Hb_002107_110 |
0.0887337735 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_092458_010 |
0.1006285632 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_005867_030 |
0.1031598581 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 5 |
Hb_007416_170 |
0.1110501066 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 6 |
Hb_001619_020 |
0.1182425491 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 7 |
Hb_000237_070 |
0.1235184902 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 8 |
Hb_002615_010 |
0.129747005 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 9 |
Hb_000177_130 |
0.1306307322 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 10 |
Hb_002010_020 |
0.1326110368 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 11 |
Hb_000495_050 |
0.133171819 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 12 |
Hb_005463_070 |
0.1357893284 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_003387_080 |
0.1358186708 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_004880_100 |
0.1375819289 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 15 |
Hb_005016_080 |
0.1381558439 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 16 |
Hb_001635_230 |
0.1384249863 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000318_050 |
0.1388125577 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000359_260 |
0.1437842274 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 19 |
Hb_005814_010 |
0.1484632651 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_002890_320 |
0.1488949266 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |