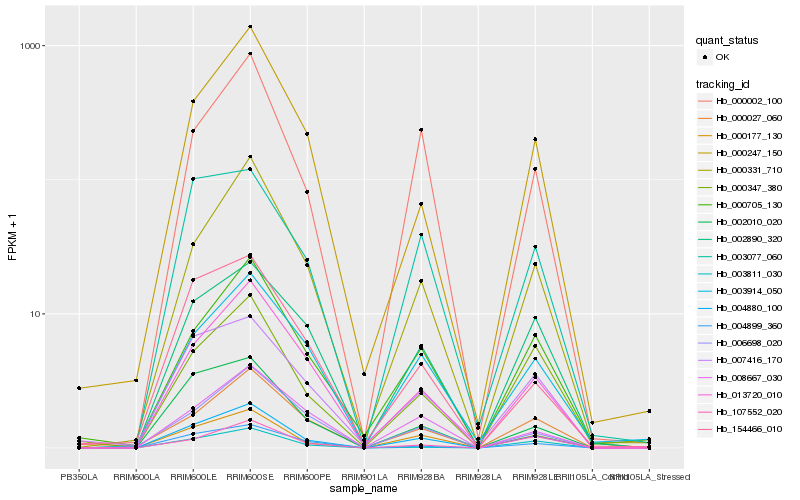
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004880_100 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |
| 2 |
Hb_000177_130 |
0.074164748 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 3 |
Hb_013720_010 |
0.116482178 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 4 |
Hb_000331_710 |
0.1209594179 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002010_020 |
0.122971678 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 6 |
Hb_000002_100 |
0.125343897 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_000705_130 |
0.1256788403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000027_060 |
0.1264156469 |
- |
- |
hypothetical protein POPTR_0004s07470g [Populus trichocarpa] |
| 9 |
Hb_002890_320 |
0.1270655065 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 10 |
Hb_007416_170 |
0.1286333076 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 11 |
Hb_003914_050 |
0.1340476277 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 12 |
Hb_004899_360 |
0.1364568053 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 13 |
Hb_003077_060 |
0.1375819289 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 14 |
Hb_000247_150 |
0.1414605199 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_000347_380 |
0.1414679982 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_107552_020 |
0.1451624935 |
- |
- |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 17 |
Hb_154466_010 |
0.1529996775 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_006698_020 |
0.1531930455 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 19 |
Hb_003811_030 |
0.1533137961 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 20 |
Hb_008667_030 |
0.1535600977 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |