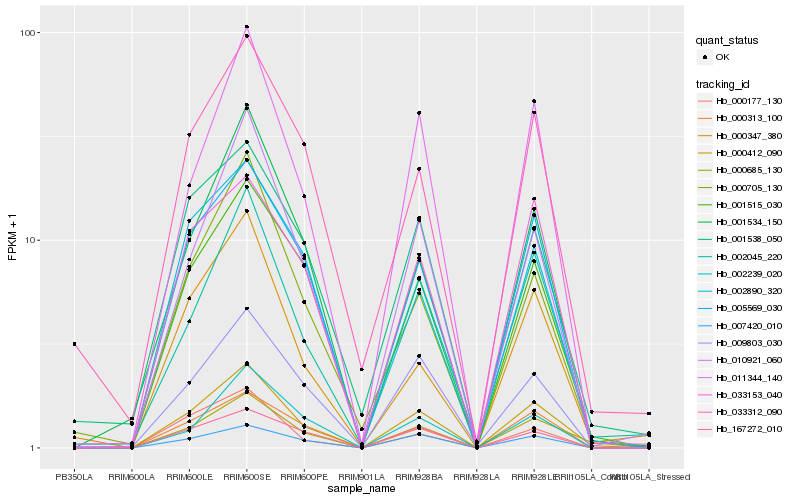
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000412_090 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_000685_130 |
0.0820511222 |
- |
- |
PREDICTED: uncharacterized protein LOC105351820 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_001515_030 |
0.1052690963 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_002045_220 |
0.1073041063 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005569_030 |
0.1077549076 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011344_140 |
0.110896416 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 7 |
Hb_007420_010 |
0.1141183868 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 8 |
Hb_010921_060 |
0.1241015854 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_033153_040 |
0.1255471494 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_009803_030 |
0.1257034459 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 11 |
Hb_167272_010 |
0.1300190459 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 12 |
Hb_000705_130 |
0.1339254919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002890_320 |
0.1354991296 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 14 |
Hb_000313_100 |
0.1383915798 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 15 |
Hb_000177_130 |
0.1385442087 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 16 |
Hb_033312_090 |
0.1398420894 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001538_050 |
0.1422448776 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 18 |
Hb_002239_020 |
0.1452899069 |
- |
- |
MLO, putative [Theobroma cacao] |
| 19 |
Hb_001534_150 |
0.1486011564 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 20 |
Hb_000347_380 |
0.1492613925 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |