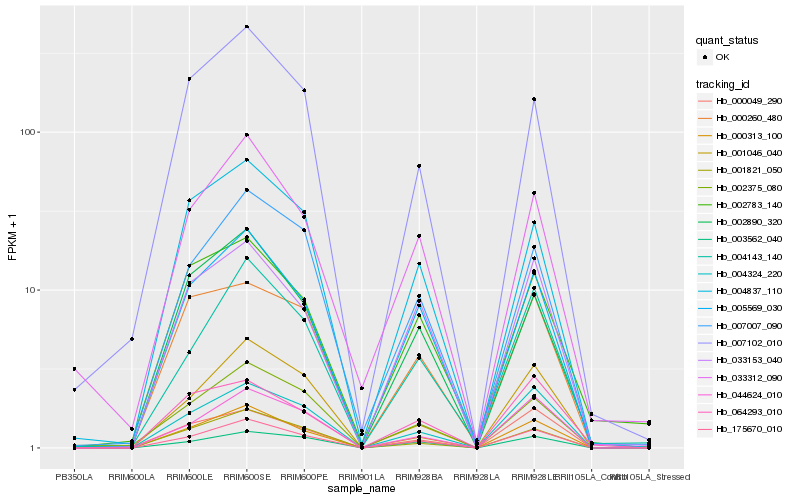
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_100 |
0.0 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 2 |
Hb_175670_010 |
0.0354549523 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 3 |
Hb_005569_030 |
0.0650992806 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 4 |
Hb_033153_040 |
0.0946103073 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_004324_220 |
0.0970957852 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 6 |
Hb_002890_320 |
0.0993156306 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 7 |
Hb_001821_050 |
0.1092864822 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 8 |
Hb_004837_110 |
0.1128611451 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_044624_010 |
0.1136092096 |
- |
- |
- |
| 10 |
Hb_003562_040 |
0.1136447091 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 11 |
Hb_004143_140 |
0.114114462 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 12 |
Hb_001046_040 |
0.1155169799 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_033312_090 |
0.121329936 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_007102_010 |
0.122412927 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 15 |
Hb_007007_090 |
0.1239933429 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 16 |
Hb_000049_290 |
0.125413557 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_002375_080 |
0.1259945613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000260_480 |
0.1282396754 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 19 |
Hb_064293_010 |
0.1313974992 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_002783_140 |
0.1326486448 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |