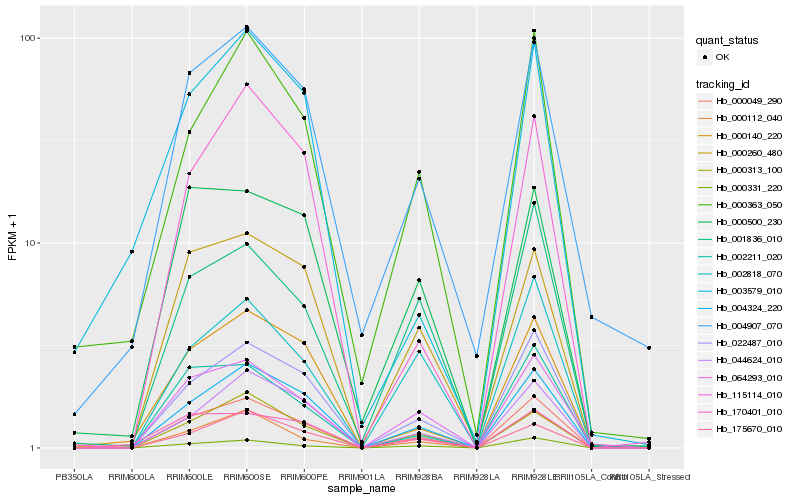
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_290 |
0.0 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_022487_010 |
0.0879644247 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 3 |
Hb_004324_220 |
0.0913479737 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 4 |
Hb_000363_050 |
0.1016936048 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000140_220 |
0.1037560527 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 6 |
Hb_064293_010 |
0.1105592285 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_000331_220 |
0.1189808987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000112_040 |
0.1204431683 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_044624_010 |
0.121639996 |
- |
- |
- |
| 10 |
Hb_000313_100 |
0.125413557 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 11 |
Hb_002211_020 |
0.1259010539 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 12 |
Hb_004907_070 |
0.1293835372 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 13 |
Hb_000260_480 |
0.1307846978 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 14 |
Hb_170401_010 |
0.1320483981 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 15 |
Hb_175670_010 |
0.1324558742 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_001836_010 |
0.1400365569 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 17 |
Hb_000500_230 |
0.1400625026 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 18 |
Hb_115114_010 |
0.1405516531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002818_070 |
0.1431027771 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 20 |
Hb_003579_010 |
0.1432028548 |
- |
- |
cryptochrome 1.2 [Populus tremula] |