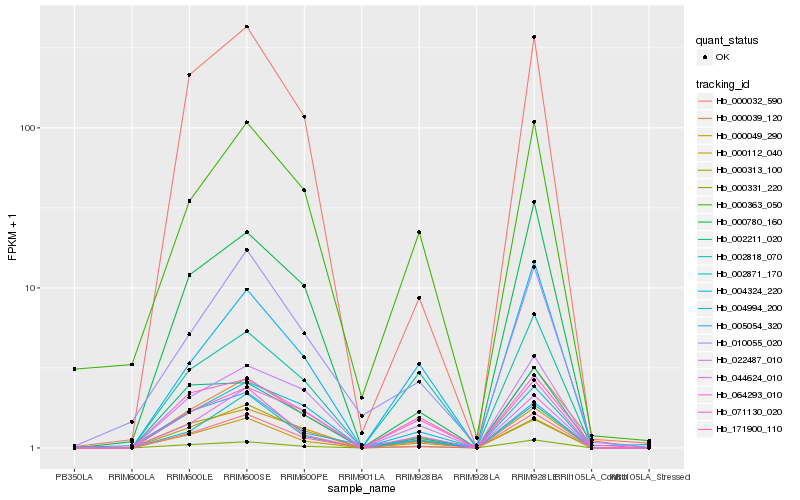
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000112_040 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_000331_220 |
0.072192572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002871_170 |
0.0811956937 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 4 |
Hb_171900_110 |
0.1043413393 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 5 |
Hb_000049_290 |
0.1204431683 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_071130_020 |
0.125309927 |
- |
- |
- |
| 7 |
Hb_000032_590 |
0.13530976 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 8 |
Hb_022487_010 |
0.1354909794 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 9 |
Hb_004994_200 |
0.1376507164 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 10 |
Hb_000363_050 |
0.138416249 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_002818_070 |
0.1401637367 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_064293_010 |
0.1410893799 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_000313_100 |
0.1417839422 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 14 |
Hb_004324_220 |
0.1421487736 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 15 |
Hb_002211_020 |
0.146184465 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 16 |
Hb_000039_120 |
0.1466792942 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 17 |
Hb_010055_020 |
0.1504943496 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 18 |
Hb_005054_320 |
0.1511422317 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 19 |
Hb_000780_160 |
0.1546877828 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 20 |
Hb_044624_010 |
0.1551191953 |
- |
- |
- |