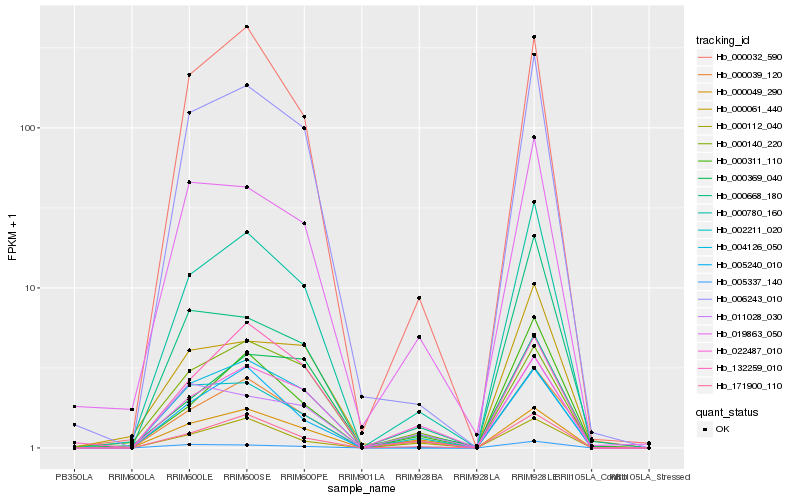
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000780_160 |
0.0 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 2 |
Hb_004126_050 |
0.0689748095 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |
| 3 |
Hb_006243_010 |
0.0802152168 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 4 |
Hb_171900_110 |
0.0898785403 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 5 |
Hb_000039_120 |
0.1165450904 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 6 |
Hb_000032_590 |
0.1214118512 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 7 |
Hb_019863_050 |
0.1241210211 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 8 |
Hb_000311_110 |
0.1243186224 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_005240_010 |
0.1262982246 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 10 |
Hb_000061_440 |
0.1271336404 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_002211_020 |
0.1282554789 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 12 |
Hb_022487_010 |
0.1328485876 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 13 |
Hb_005337_140 |
0.138013691 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase 1-like [Jatropha curcas] |
| 14 |
Hb_000049_290 |
0.1501459417 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_000668_180 |
0.1516160639 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 16 |
Hb_132259_010 |
0.1518218525 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 17 |
Hb_000140_220 |
0.1530547839 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 18 |
Hb_011028_030 |
0.1538822088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000112_040 |
0.1546877828 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_000369_040 |
0.1547449928 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |