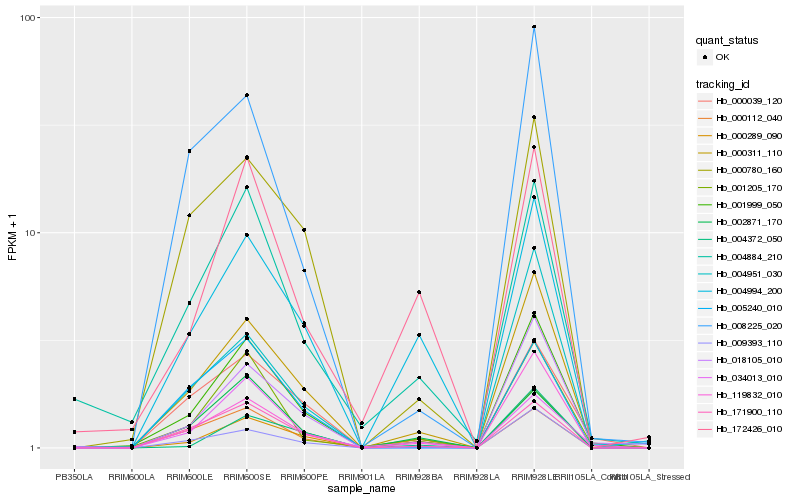
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_050 |
0.0 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 2 |
Hb_000311_110 |
0.0793307907 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 3 |
Hb_018105_010 |
0.1146749129 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 4 |
Hb_001205_170 |
0.1280427046 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 5 |
Hb_119832_010 |
0.135487724 |
- |
- |
- |
| 6 |
Hb_000039_120 |
0.143439546 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 7 |
Hb_171900_110 |
0.1501229354 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 8 |
Hb_009393_110 |
0.1507466685 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 9 |
Hb_004884_210 |
0.155343052 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 10 |
Hb_000780_160 |
0.1574324067 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 11 |
Hb_008225_020 |
0.1626780868 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 12 |
Hb_004994_200 |
0.1724715483 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 13 |
Hb_034013_010 |
0.1753871726 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 14 |
Hb_000289_090 |
0.1756465193 |
- |
- |
- |
| 15 |
Hb_004372_050 |
0.1811903627 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 16 |
Hb_005240_010 |
0.1846768365 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 17 |
Hb_002871_170 |
0.1859358584 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 18 |
Hb_004951_030 |
0.1872196885 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_000112_040 |
0.1874582075 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_172426_010 |
0.1888410991 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |