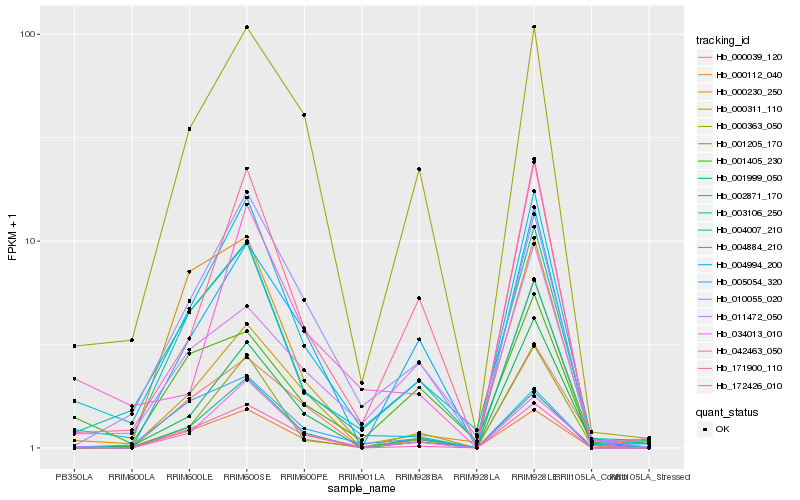
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004884_210 |
0.0 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 2 |
Hb_003106_250 |
0.1108592831 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 3 |
Hb_034013_010 |
0.1372274375 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 4 |
Hb_010055_020 |
0.1456120042 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 5 |
Hb_172426_010 |
0.1487179806 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 6 |
Hb_001205_170 |
0.1500790459 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000363_050 |
0.153781875 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001999_050 |
0.155343052 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 9 |
Hb_002871_170 |
0.1615977051 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 10 |
Hb_000112_040 |
0.1636592436 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_000311_110 |
0.1648059982 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 12 |
Hb_171900_110 |
0.169928076 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 13 |
Hb_004007_210 |
0.1721267141 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 14 |
Hb_004994_200 |
0.1825629425 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 15 |
Hb_005054_320 |
0.1854982405 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 16 |
Hb_042463_050 |
0.1856421787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001405_230 |
0.1893328022 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 18 |
Hb_000039_120 |
0.189785344 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 19 |
Hb_000230_250 |
0.1914990985 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_011472_050 |
0.1933626703 |
- |
- |
- |