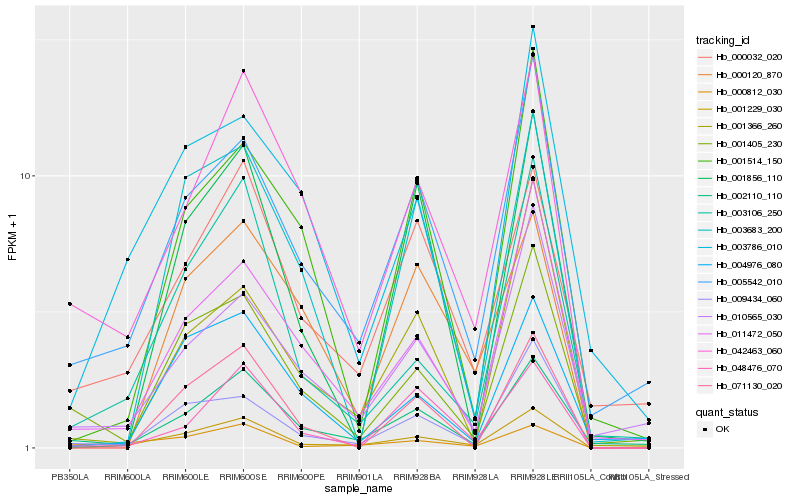
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001229_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104783713 [Camelina sativa] |
| 2 |
Hb_003106_250 |
0.1498547843 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 3 |
Hb_009434_060 |
0.1636007643 |
- |
- |
T4.15 [Malus x robusta] |
| 4 |
Hb_005542_010 |
0.1674907749 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 5 |
Hb_011472_050 |
0.1735893843 |
- |
- |
- |
| 6 |
Hb_002110_110 |
0.1768676067 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 7 |
Hb_042463_060 |
0.1839585292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000812_030 |
0.1877458626 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 9 |
Hb_010565_030 |
0.194459104 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 10 |
Hb_000032_020 |
0.195846325 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 11 |
Hb_004976_080 |
0.1970698116 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 12 |
Hb_071130_020 |
0.1974233271 |
- |
- |
- |
| 13 |
Hb_048476_070 |
0.1980431017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001405_230 |
0.2009629844 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 15 |
Hb_001366_260 |
0.2032624941 |
- |
- |
T4.15 [Malus x robusta] |
| 16 |
Hb_003786_010 |
0.2049481632 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003683_200 |
0.2050030282 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 18 |
Hb_000120_870 |
0.2074732275 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 19 |
Hb_001514_150 |
0.2078630884 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 20 |
Hb_001856_110 |
0.2097903018 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |