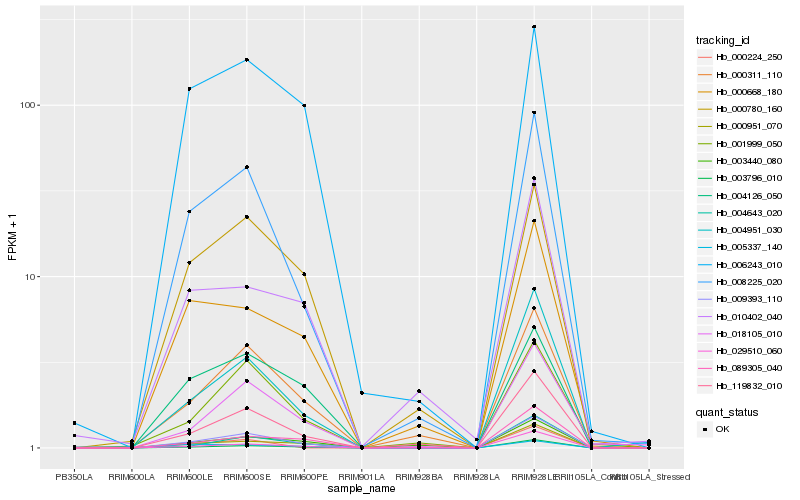
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009393_110 |
0.0 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 2 |
Hb_003796_010 |
0.0829213999 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_004951_030 |
0.0848929782 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_008225_020 |
0.0996681029 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 5 |
Hb_119832_010 |
0.1004883573 |
- |
- |
- |
| 6 |
Hb_000311_110 |
0.1215522247 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 7 |
Hb_000951_070 |
0.1258626646 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 8 |
Hb_004643_020 |
0.1278622503 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 9 |
Hb_018105_010 |
0.1284149257 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 10 |
Hb_089305_040 |
0.1316910343 |
- |
- |
- |
| 11 |
Hb_001999_050 |
0.1507466685 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 12 |
Hb_000668_180 |
0.1580973534 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 13 |
Hb_000780_160 |
0.1586766313 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 14 |
Hb_004126_050 |
0.1647182614 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |
| 15 |
Hb_029510_060 |
0.1689296635 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 16 |
Hb_003440_080 |
0.1703478773 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 17 |
Hb_010402_040 |
0.1767148265 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 18 |
Hb_005337_140 |
0.1804327242 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase 1-like [Jatropha curcas] |
| 19 |
Hb_000224_250 |
0.1843952229 |
- |
- |
hypothetical protein AMTR_s05400p00003230 [Amborella trichopoda] |
| 20 |
Hb_006243_010 |
0.185759141 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |