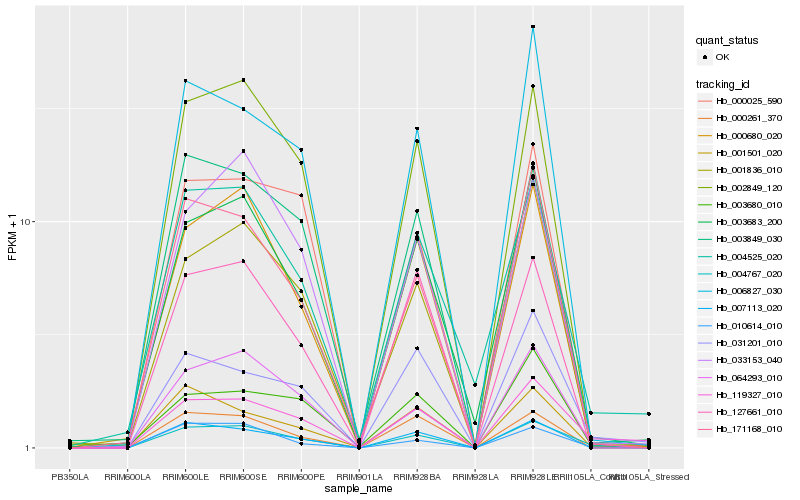
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004767_020 |
0.0 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_064293_010 |
0.1115774993 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 3 |
Hb_119327_010 |
0.1227590962 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 4 |
Hb_002849_120 |
0.1327505088 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 5 |
Hb_171168_010 |
0.1397644046 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_007113_020 |
0.1400075717 |
- |
- |
- |
| 7 |
Hb_000680_020 |
0.1408519926 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_003849_030 |
0.1412665725 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 9 |
Hb_010614_010 |
0.1415336044 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 10 |
Hb_003683_200 |
0.1423458127 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 11 |
Hb_004525_020 |
0.1423908643 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 12 |
Hb_033153_040 |
0.1519592058 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_006827_030 |
0.1531565946 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 14 |
Hb_127661_010 |
0.1558908413 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_001501_020 |
0.1564500815 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 16 |
Hb_031201_010 |
0.1568500574 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000025_590 |
0.159133791 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001836_010 |
0.1592288921 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 19 |
Hb_003680_010 |
0.1592936439 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000261_370 |
0.1613995693 |
- |
- |
- |