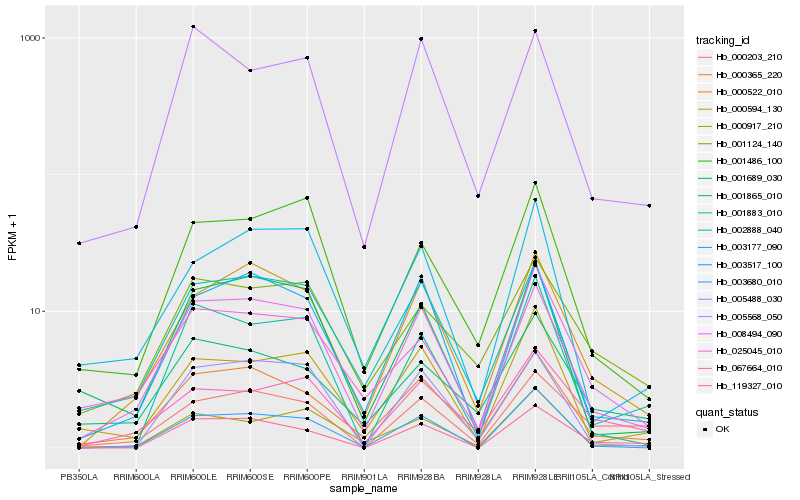
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008494_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 2 |
Hb_000365_220 |
0.0850926834 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 3 |
Hb_000522_010 |
0.0869065421 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 4 |
Hb_025045_010 |
0.1279383688 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_003680_010 |
0.1294055542 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_067664_010 |
0.1296114116 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 7 |
Hb_002888_040 |
0.1303036699 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 8 |
Hb_001486_100 |
0.1310904688 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 9 |
Hb_003517_100 |
0.131730627 |
transcription factor |
TF Family: GRAS |
- |
| 10 |
Hb_000917_210 |
0.1322096283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001689_030 |
0.1347703495 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 12 |
Hb_000594_130 |
0.140461012 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_119327_010 |
0.1421729192 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 14 |
Hb_000203_210 |
0.1429626625 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 15 |
Hb_003177_090 |
0.1450697051 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 16 |
Hb_001124_140 |
0.1450952124 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 17 |
Hb_001883_010 |
0.147237292 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 18 |
Hb_001865_010 |
0.1479827361 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 19 |
Hb_005488_030 |
0.148354648 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 20 |
Hb_005568_050 |
0.1488254399 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |